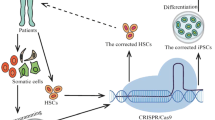
Abstract
Rapid advancement in genome editing technologies has provided new promises for treating neoplasia, cardiovascular, neurodegenerative, and monogenic disorders. Recently, the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) system has emerged as a powerful gene editing tool offering advantages, including high editing efficiency and low cost over the conventional approaches. Human pluripotent stem cells (hPSCs), with their great proliferation and differentiation potential into different cell types, have been exploited in stem cell-based therapy. The potential of hPSCs and the capabilities of CRISPR/Cas9 genome editing has been paradigm-shifting in medical genetics for over two decades. Since hPSCs are categorized as hard-to-transfect cells, there is a critical demand to develop an appropriate and effective approach for CRISPR/Cas9 delivery into these cells. This review focuses on various strategies for CRISPR/Cas9 delivery in stem cells.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Genome Editing and Programmable Nucleases
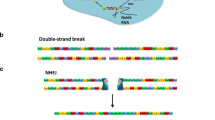
In recent years, advances in delivery technologies have enabled the intracellular loading of impermeable gene-editing tools, such as programmable nucleases, into different cell types. These technologies, coined as genome editing approaches, can lead to alterations in genomic DNA sequence through either insertion or deletion (indels) of one or more base pairs. Genome editing approaches have been exploited in various kinds of diseases, from treating life-threatening conditions (e.g., hereditary disorders) to restoring the lost function in gene expression studies [1, 2]. The genome-editing procedure mainly relies on creating double-strand breaks (DSBs) at the site of the target sequence. These site-specific DSBs can be repaired through two distinct endogenous pathways: i) non-homologous end-joining (NHEJ), which is the primary repairing pathway throughout the cell cycle, and ii) homology-directed repair (HDR). In NHEJ, protein factors force the DSBs to re-join and ligate the cleaved DNA strands without requiring a homologous template leading to the formation of indels followed by modification or inactivation of the target gene. With lower frequency, the HDR pathway can repair the DSBs only in the presence of an appropriate DNA donor template [3].
To date, different types of engineered nucleases, including meganucleases, transcription activator-like effector nucleases (TALENs), zinc-finger nucleases (ZFNs), CRISPR/Cas9, and prime editing complexes have been developed to generate DSBs at targeted DNA sequences [4] (Fig. 1). These engineered nucleases recognize the target sites through interactions of DNA-protein (meganucleases, TALENs, and ZFNs) or DNA-RNA (CRISPR/Cas9).
Schematic of the gene-editing procedure. Sample preparation process (I) and the leading gene-editing platforms, including ZFNs (II), CRISPR/Cas9 complexes (III), and prime editing complexes (IV). Sample preparation involves isolating stem cells from the patient blood sample, followed by expanding the isolated cells in culture. Next, ex vivo genetic engineering will take place, which involves creating double-strand breaks in DNA using programmable nucleases, CRISPR/Cas9 complex, or prime editing system
CRISPR/Cas9 is a novel gene-editing approach derived from the prokaryotic immune system produced in response to the exogenous nucleic acids of phages and plasmids. Since 2013, when Cong et al. employed CRISPR/Cas technology for efficient genome editing in eukaryotic cells, this approach has gained significant attention in the field [5]. Moreover, CRISPR/Cas editing tool has shown many advantages over the other engineered nucleases, such as i) guiding the nuclease to the targeted site through the simple base-pairing rules, ii) the possibility of synthesizing the guiding RNAs in vitro, and iii) generation of multiple DSBs synchronously by using different guide RNA sequences, which enables editing of several loci in the mammalian genome. Recent studies have employed CRISPR-Cas9 as the most popular gene editing approach to study cystic fibrosis [6, 7], sickle cell disease (SCD) [8, 9], Huntington’s chorea disorder [10, 11], Duchenne muscular dystrophy [12,13,14], chronic granulomatous disease [15, 16] retinitis pigmentosa [17] hemophilia [18, 19], and thalassemia [20,21,22].
The CRISPR/Cas9 system mainly comprises a nuclease, called Cas9, and a single guide RNA (sgRNA) containing a sequence-specific CRISPR RNA (crRNA), capable of recognizing 18–20 nucleotides of the target DNA, and an auxiliary trans-activating crRNA (tracrRNA) [23]. Currently, different systems are used in CRISPR/Cas9 mediated gene editing, including plasmid-based CRISPR/Cas9, ribonucleoprotein (RNP) complex-based Cas9 protein with sgRNA, and Cas9 mRNA with sgRNA [24].
In plasmid-based CRISPR/Cas9 gene editing, plasmid DNA encoding both customized sgRNA and Cas9 protein enters the nucleus to be transcribed into target sgRNA and Cas9 mRNA. Next, the Cas9 transcript is translated into Cas9 protein in the cytoplasm and then returned to the nucleus for genome editing [25]. Although CRISPR/Cas9 plasmid DNAs are more stable than CRISPR/Cas9 mRNA and RNP complexes, potential disadvantages attributed to this approach are the long-lasting expression of the CRISPR/Cas9 system, increased off-target effects, and the possibility of DNA integration into the host genome [26]. On the contrary, in mRNA-based CRISPR/Cas9 gene editing, the transcription of sgRNA and Cas9 mRNA is not required. Therefore, efficient gene editing can be performed faster while reducing the off-target effects. Upon cytoplasmic delivery of the sgRNA and Cas9 mRNA, the mRNA translation would take place to produce the Cas9 protein, followed by localization of both sgRNA and Cas9 protein to the nucleus for gene editing [27].
The RNP complex-based system (Cas9 protein and sgRNA) is the most efficient and rapid gene editing approach, in which neither transcription nor translation is required. Since the CRISPR/Cas9 RNP complex does not naturally exist in the cells of interest, this highly efficient approach offers minimal off-target effects. Despite these advantages, this approach suffers from the high cost of producing RNP complex and toxic side effects [28].
Stem Cell-Based Therapeutic Applications of CRISPR
A variety of genetic disorders arise from mutations at the genomic level. Stem cell-based therapies have demonstrated efficacy in treating these diseases in clinical trials. This promising strategy contains the autologous transplantation of genetically edited stem cells. These genetically modified stem cells offer several advantages, such as self-renewal capacity, increasing the patient’s lifetime, and dividing into daughter cells with the ability to be differentiated into resident cells within different tissues [29]. Growing evidence indicated that genome editing through CRISPR/Cas9 system is an efficient approach extending to stem cell-based research and therapies. HPSCs have great proliferation potential and can differentiate into various cell types. These cells (e.g., neural, hematopoietic, and mesenchymal stem cells) can be manipulated using CRISPR/Cas9 gene editing tools and employed for therapeutic applications [30, 31]. An online search ranging from 2010 until 2022 (15/07/22) was performed on PubMed using the terms “CRISPR/Cas9” and “stem cell”. The search results revealed that several studies used the CRISPR/Cas9 system for the genetic engineering of the hPSCs (Fig. 2). Since the hPSCs are naturally impermanent to external cargoes such as CRISPR/Cas9 gene editing tools, these studies have used external forces for transient cell membrane disruption and cytoplasmic loading of these tools inside the cells, which will be discussed in the next section.
Research studies on the therapeutic applications of CRISPR/Cas9-mediated gene editing in stem cells. I) Since 2013, several delivery technologies have been used for cytoplasmic delivery of the CRISPR/Cas9 gene editing tools into the hPSCs. Different delivery techniques (II) have been used to load various cargoes into a wide range of cell types (III)
Delivery Systems for Gene Editing by CRISPR/Cas9 in Stem Cells
During the past decades, several delivery technologies have been developed to transiently permeabilize hPSCs plasma membrane, followed by facilitating the cytoplasmic delivery of the CRISPR/Cas9 genome editing tools. These technologies have successfully loaded the CRISPR/Cas9 tools inside the hard-to-transfect hPSCs for efficient genome editing with high specificity delivery efficiency [32]. This review focuses on carrier independent (e.g., physical and mechanical delivery) (Fig. 3) and dependent (e.g., nanoparticle, extracellular vesicles, viral-like particles, and viruses) strategies that have been employed to load CRISPR/Cas9 gene editing tools into different types of stem cells (Fig. 4).
Carrier independent strategies for CRISPR/Cas9 delivery into stem cells. These approaches can be divided into physical methods, including electroporation and induced transduction by osmosis and propanebetaine (iTOP), and mechanical methods, such as microfluidic-, silicon nanoblade-, and transmembrane internalization assisted by membrane filtration (TRIAMF)-based delivery systems
Carrier Independent Delivery Approaches
Carrier-independent approaches comprise physical and mechanical methods that utilize external forces to concurrently induce transient membrane disruptions and cargo delivery. These modalities, also coined as direct membrane permeabilization strategies, can create pores of different sizes allowing the passive diffusion of exogenous cargoes across the plasma membrane [33]. In this section, we will cover physical and mechanical delivery approaches that have been exploited for CRISPR/Cas delivery into the cells of interest.
Physical Delivery
Physical delivery methods are considered an effective approach for the direct delivery of nucleic acids, also called transfection, into the hard-to-transfect cells [34]. During the past decade, physical delivery approaches have been widely used for loading both CRISPR/Cas9 plasmid DNA and RNP into various cell types. The commonly used approaches for delivering CRISPR/Cas9 into different types of stem cells include electroporation, nucleofection, induced transduction by osmocytosis and propanebetaine (iTOP) [35], and mechanical transfection [36], which will be covered in the following sections. Although these approaches are simple and highly reproducible, challenges include processing a bulk population of cells resulting in heterogeneous responses within the cell population [37, 38].
Electroporation
Electroporation is a physical delivery approach that uses a series of controlled electric pulses to induce transient membrane permeabilization and cytosolic uptake of impermeable macromolecules such as plasmid DNA and gene constructs [39]. In this approach, a bulk population of cells is resuspended in an electroconductive buffer and added to a cuvette placed between two electrodes. Therefore, applying controlled electric pulses with optimized voltages and widths to the target cells can result in the creation of transient pores across the plasma membrane, facilitating the passive influx of the impermeable plasmid DNA [32]. Electroporation is considered as a highly efficient delivery approach that can increase the overall uptake and expression level of the exogenous DNA up to 1000 folds. It is worth noting that the delivery efficiency of this technique mainly relies on different parameters, including electric field characteristics (voltage, width, and number of pulses), electrode geometry, and cell and cargo types. One-size-fits-all gene delivery through electroporation has been elusive as optimal delivery conditions must be determined based on different cell and cargo types. Gene delivery using cuvette-based electroporation has shown promise in different applications, from vaccine development, transgenes expression, and enzyme replacement to cancer treatment. Since cuvette-based electroporation is a heterogeneous treatment that can induce different levels of stress in the target cells depending on their position relative to the electrodes, the clinical application of this conventional approach has been restricted [40]. Besides this, electroporation is a costly method that requires rather expensive reagents and kits, hindering its application in many laboratories.
Along with the technological advances in the field, significant efforts have been made toward implementing electroporation at the microscale. As a result, the Neon™ transfection system, which works by inducing an electric field within capillaries, has been developed and further commercialized by Invitrogen/Thermo fisher. This new user-friendly setup is more flexible and is designed to address the challenges of the conventional cuvette-based electroporation strategy. In this setup, the transfection will happen within a biologically compatible pipette tip chamber, which has been employed to generate more uniform electric fields. This technique maximizes the distance between electrodes while minimizing their surface area to ensure a controlled electric pulse is passed through the target cells to achieve a homogenous exposure and treatment. This rapid and highly reproducible technique has significantly improved the delivery outcomes in hard-to-transfect cells compared to the cuvette-based electroporation [41]. In 2019, this technology was used for the co-delivery of Cas9 protein, gRNA, and two indistinguishable donor DNA templates into induced pluripotent stem cells (iPSCs). This resulted in increased efficiency (~8.3%) of inducing in vitro homozygous modifications while maintaining cell viability [42].
In 2018, Xu et al. reported a tube-based electroporation method for efficient genome editing by delivering the CRISPR/Cas9 RNP into nearly 90% of the iPSCs without affecting the cell viability. As a result, a relatively high HDR rate (42.1%) was achieved in the edited cells, which is significantly higher than those reported in the previous studies within the range of 2.1–6.7% [43, 44]. In a recent study, Yudovich and colleagues performed viral-mediated delivery of sgRNAs into hematopoietic stem and progenitor cells (HSPCs), followed by Cas9 mRNA delivery using BTX ECM 830 electroporation. Using this combinational strategy, they could achieve up to 90% knockout of CD44 and CD45 cell surface proteins [45].
Nucleofection is another commercially available gene delivery strategy, which is mostly designed for the nuclear delivery of cargoes like siRNA, DNA, and oligonucleotides into hard-to-transfect and primary cell lines. This method is one of the most popular cuvette-based electroporation configurations that utilizes cell line-specific buffers along with nucleofector pulsing parameters for the direct delivery of nucleic acids into the nucleus. Nucleofection has become a popular protocol for the highly efficient delivery of CRISPR/Cas9 plasmids into different types of stem cells regardless of the cell cycle status [46,47,48]. Using the Amaxa Nucleofector 2 device (Lonza), Shinkuma et al. delivered CRISPR/Cas9 plasmid into the iPSCs to target a dominant negative mutation (c.8068_8084delinsGA) of COL7A1 gene in dominant dystrophic epidermolysis bullosa (DDEB). Using this strategy, they could achieve up to 90% editing efficiency through the NHEJ pathway in iPSCs generated from the DDEB patient’s fibroblasts [49]. In a follow-up study, Amaxa Nucleofector 2 device was used for co-transfection of spyFiCas9 and gRNA expression vectors into the iPSCs of recessive dystrophic epidermolysis bullosa (RDEB) patients. As a result, single and double allele mutation corrections in COL7A1 were achieved by activating the HDR pathway with efficiencies of 40% and 10%, respectively [50]. While HDR-mediated gene correction results in lower efficiencies, it is considered to be a more precise gene editing pathway than NHEJ. The lower efficiencies achieved through the HDR pathway can be attributed to the lower frequency of homologous recombination in target cells. Table 1 summarizes the studies that leveraged electroporation for CRISPR/Cas9 delivery, mostly in the form of CRISPR/Cas9 RNP, into different types of stem cells [136,137,138].
Induced Transduction by Osmocytosis and Propanebetaine (iTOP)
iTOP is another carrier independent delivery strategy that utilizes a hyperosmolar buffer to load cargoes, such as proteins, into different cell types. This delivery buffer consists of propanebetaine and sodium chloride acting as a transduction compound to trigger macropinocytosis and non-specific internalization of the extracellular fluid through engulfment of the 0.5–5 μm vesicles. This approach has been used for efficient intracellular delivery of sgRNAs and Cas9 protein separately and together in the form of CRISPR/Cas9 RNP complex [139]. The iTOP method allows the loaded protein to transiently manipulate the cells and induce changes in cell function or epigenetic status. However, this approach has not gained much attraction due to its lower gene editing efficiency in primary cells compared with other commonly used delivery technologies [34, 140,141,142,143]. D’Astolfo and colleagues performed one and two consecutive rounds of iTOP to deliver CRISPR/Cas9 to human embryonic stem cells while achieving 10% and 26% gene editing efficiency, respectively [141]. Since iTOP leverages NaCl-mediated delivery of the Cas9 protein, it can induce damage to desired cells and delivery protein resuspended in a highly concentrated salty solution, making this strategy inapplicable for in vivo gene editing purposes.
Mechanical Delivery
Creating transient ruptures in the phospholipid bilayer can be achieved by inducing mechanical forces to the cells of interest. Mechanical delivery, coined as mechanoporation, has been performed through physical contact with a firm structure or exposure to fluid shear forces directed at the cell surface. These mechanisms will result in lipid heads’ instabilities, facilitating the transient pore formation and passive cargo transports [144]. One advantage of mechanical delivery is the efficient delivery of large-sized cargoes, including plasmid DNA, which is challenging using benchtop delivery options. Moreover, this approach has shown utility for the efficient delivery of genetic materials into hard-to-transfect immune and stem cells. In recent studies, mechanical delivery has been employed for CRISPR/Cas9 RNP complex delivery into HSPCs demonstrating the further applicability for ex vivo cell therapies [145, 146]. In 2017, Ma et al. developed a microfluidic chip named nano-blade chip (NB-Chip) for transient mechanical deformation and highly efficient delivery of the CRISPR/Cas9 complex into the CD34+ HSPCs. The proposed asymmetrical microchannel features a silicon nanoblade structure on one side of the deformation zone to induce contact pressure to the CD34+ HSPCs leading to membrane disruption. During the design optimization, the nano-blade structure stiffness and sharpness were substantially increased, which further assisted the efficient delivery of biomaterials. Furthermore, the treated HSPCs had long-term viability and maintained inherent multi-potency [145].
Another form of mechanical delivery is filtroporation which utilizes fluid shear forces to generate disruptions in the plasma membrane. This technique gained less attention until 2018 when transmembrane internalization assisted by membrane filtration (TRIAMF) was developed based on the filtroporation using track-etched membranes to load CRISPR/Cas9 complex targeting β2-microglobulin into CD34+ HPSCs. By forcing the cell suspension through the membrane’s micropores, fluid shear forces are generated, which further induce cellular stress to the point of creating transient membrane ruptures and, in turn, intracellular loading of cargo molecules to the target cells [146].
Carrier Dependent Delivery Approaches
Carrier dependent delivery strategies rely on biological and chemical vectors to encapsulate the exogenous cargoes and bypass the plasma membrane barrier. Not only do these vectors protect the cargo from degradation but also, they facilitate the internalization of the cargo to the intended intracellular compartment. The cargo internalization mechanism for vectors is either through endocytosis or membrane fusion. This section will provide further details on different carrier dependent delivery systems, including nanoparticles, extracellular vesicles, virus-like particles, and viral vectors (Fig. 4).
Nanoparticle Delivery
In recent years, nanoparticles (NPs) have been widely used as promising carriers of the CRISPR/Cas9 complex, attracting great attention. Remarkable advances in nanoparticle research have revolutionized the field of controlled therapeutic delivery due to the advantages, including high efficiency, low cost, non-immunogenicity, and non-mutagenicity. To date, different types of lipid- and polymer-based as well as gold NPs have been developed and used for CRISPR/Cas9 delivery into different cell types [147]. In the following sections, we will provide an overview of the mechanism of action and application of these NPs in CRISPR/Cas9 delivery research.
Lipid-Based NPs (LNPs)
Lipid-based NPs, also known as liposomes, are spherical vesicles made up of phospholipid bilayers acting as effective delivery vehicles in different biological applications, including intracellular delivery of genetic materials into the target cells and treatment of different diseases in clinical practice [148]. Since plasma membrane and nucleic acids both feature negative charges, the electrostatic repulsion hinders the entrance of exogenous nucleic acids into the cells of interest. To overcome this challenge, nucleic acids are encapsulated into positively charged liposomes facilitating the cargo delivery and subsequent cellular uptake by converting the repulsive to attractive electrostatic forces [149]. LNP-mediated CRISPR/Cas9 delivery is a food and drug administration (FDA) approved strategy, which induces less stress to the desired cells leading to higher cell viability than its counterparts. However, this modality suffers from low delivery efficiency as it mainly relies on the endosomal pathway for cargo internalization [150, 151].
Advances in nanotechnology have enabled the development of the lipofectamine reagent, which is now the first preferred option for LNP-mediated delivery [150]. In a study, Lipofectamine® 3000 has been used as a CRISPR/Cas9 delivery tool for correction of the suspected causative SCN5A variant (rs397514446) in iPSCs-derived cardiomyocytes of patients with Brugada syndrome (BrS) [152]. Further studies with a focus on gene editing through lipofectamine-mediated delivery of CRISPR/Cas9 system into stem cells are summarized in Table 2.
Polymer-Based NPs
In recent studies, cationic polymer-based NPs have been widely used for different purposes, including gene delivery. These NPs can form polyplexes containing nucleic acids (nucleic acid/polycation complexes) through electrostatic interactions between the cationic group of the NPs and negatively charged nucleic acids [169, 170]. Polymer-based NPs can be formulated with different copolymer compositions and molecular weights with various degradation times ranging between several months to years [171, 172]. These NPs have been reported to improve the carrier-mediated delivery of CRISPR/Cas9 components [173]. Among the polymer-based NPs employed for CRISPR/Cas9 delivery is poly lactic-co-glycolic acid (PLGA) which is a biodegradable polymer as its hydrolysis can result in the formation of glycolic acid, lactic acid, and metabolite monomers. Since glycolic and lactic acids are both endogenous and can be readily metabolized through the Krebs cycle inside the human body, using PLGA-based NPs for biological applications and drug delivery purposes has shown minimal cytotoxic effects [174]. Owing to the advantages of PLGA-based NPs, using these NPs for drug delivery applications into the human body has been approved by the US FDA and the European medicine agency (EMA).
In a recent study, PLGA-based NPs were designed to encapsulate Cas9 protein (S. pyogenes) and gRNA and used as a carrier for CRISPR/Cas9 delivery into HSPCs. Upon successful delivery of the CRISPR/Cas9 complex, a rapid release of gRNA and Cas9 protein was observed, which was subsequently replaced with a continuous cargo release pattern resulting from endosomal/lysosomal escape and cytosolic penetration. More importantly, Cruz et al. demonstrated that PLGA-based NP-mediated gene-editing of HSPCs using CRISPR/Cas9 complex did not induce cellular cytotoxicity. Upon escaping from the lysosomal compartments, CRISPR/Cas9-PLGA-based NPs could efficiently (up to 40%) edit the γ-globin gene locus resulting in a significant increase in the expression level of fetal hemoglobin in primary erythroid cells [175]. Besides the advantages offered by these NPs, size dissimilarities and unpredictable behavior and interaction of NPs with target cells are the remaining challenges of the field which require further investigations.
Gold NPs (au NPs)
Along with the advances in nanotechnologies, inorganic NPs like Au NPs and magnetic NPs were developed and employed as appropriate carriers for gene delivery applications. Among inorganic NPs, Au NPs hold the potential to be used as multifunctional gene delivery systems due to their simple synthesis and modification process, high loading capacity, high cellular uptake, and inherent biocompatibility [176, 177]. Since these NPs are chemically inert, Au NP-mediated cargo delivery usually does not induce adverse immune responses inside the body [178]. Despite these advantages, Au NPs can induce cytotoxic effects at high concentrations eliminating their applications in clinical settings. However, a large number of studies have reported the use of Au NPs for CRISPR/Cas9 RNP complex delivery in both in vivo and in vitro conditions [155, 177, 179]. In a study, Au-NP-CRISPR/Cas9 carriers were generated through multiple formulation steps. First, Au NPs with the size of 15 nm were conjugated to single-stranded DNA sequences (50 thiols modified) that were hybridized to single-stranded donor DNA and generated the Au-NP-Donor complex. Then, CRISPR/Cas9 RNPs were loaded on Au-NP-Donor complex and further coated with silica and PAsp (DET) polymer to escape the endosomal entrapment resulting in cytosolic cargo release. The resulting Au-NP-Donor-CRISPR/Cas9-silicate carriers could successfully induce HDR in primary cells and cell lines in vitro with an editing efficiency of about 4% which is higher than lipofectamine-mediated transfection and nucleofection. Furthermore, muscular injection of NP-Donor-CRISPR/Cas9-silicate carriers in four-week-old mdx mice resulted in vivo correction of a point mutation in the dystrophin gene through the HDR pathway with editing efficiency of about 5.4% [155].
In another study, a CRISPR/Cas9 delivery system was developed by nano-formulation of colloidal Au NPs with the ability to enter the cells without the aid of external forces. The final monodispersed Au NP/crRNA nanoconjugates avoided lysosomal entrapment and were localized in the nucleus of HSPCs without inducing cytotoxicity. Genome editing at different points of interest in HSPCs was successfully achieved using these NPs nano-conjugated with different CRISPR nucleases (Cpf1 or Cas9). The primary cells of humanized mice treated with the monodispersed Au NP/crRNA nanoconjugates demonstrated better engraftment kinetics compared with the untreated cells without any significant difference in differentiation [156]. Further studies are required to generate the Au NPs that can perform targeted delivery of the CRISPR/Cas9 system into the cells of interest in vivo.
Extracellular Vehicles (EVs)
EVs are cell-derived membranous structures that serve as carriers for the delivery of various types of therapeutic cargoes, such as proteins, lipids, effector molecules, and receptors, to the target cells [180]. EVs are generated through cellular activation or stress. EVs serve as communicating agents between the cells by transporting the contents and surface proteins of the parent to the target cells. Various diagnostic and discovery applications have been demonstrated for EVs. Moreover, EVs are considered promising carriers for safe and robust cell and gene therapy applications that require strong target specificities [181]. In recent years, many studies have reported highly efficient delivery of CRISPR/Cas9 RNPs both in vitro and in vivo via different types of EVs [154, 182, 183]. These studies demonstrated the capability of EVs to be used in clinical settings. EVs are categorized into microvesicles, apoptotic bodies, and exosomes based on their intracellular origins. Among these, exosomes have attracted great interest to be used as carriers for delivery purposes [184]. Exosomes are small membrane-bound EVs (30–150 nm) secreted by the cells through the endosomal route. Exosomes may contain a complex cargo of contents associated with pathological and biological situations of the original cell. Owing to the advantages of exosomes, like inherent non-immunogenicity, these therapeutic carriers have been preferred over the other nano-sized delivery carriers, like polymer-based and Au NPs as well as liposomes [185].
In a recent study, Lin and colleagues developed exosome-liposome hybrid nanocarriers to surmount the inherent limitation of exosomes in the encapsulation of large nucleic acids [158]. Unlike exosomes, these hybrid nanocarriers could successfully encapsulate large-sized plasmids like CRISPR/Cas9 expressing vectors. These hybrid nanocarriers can enter the cells through the endocytosis pathway and induce the expression of the encapsulated genes in the mesenchymal stem cells that were not efficiently transfected with liposomes. Since the exosome–liposome hybrid NPs were able to efficiently deliver CRISPR/Cas9 into mesenchymal stem cells, they can be new candidates for in vivo gene therapy applications which remain open for further investigations.
In another investigation, a new EV-based delivery technology called NanoMEDIC was developed by employing two different homing mechanisms to package CRISPR/Cas9 RNP complex. In this delivery system, Cas9 protein was recruited to extracellular nanovesicles through ligand-dependent dimerization. Next, the two self-cleaving riboswitches and a viral RNA packaging signal held and released sgRNA into these nanovesicles. The NanoMEDIC technology demonstrated >90% efficiency in exon 45 skipping in the dystrophin gene of skeletal muscle cells that were derived from iPSCs of a patient with Duchenne muscular dystrophy. Moreover, the muscular injection of these nanovesicles in mdx mice and a luciferase reporter mouse model successfully induced transient genomic exon 23 skipping in the dystrophin gene [154]. These findings suggest the potential in vivo application of NanoMEDIC for in vivo gene therapy of Duchenne muscular dystrophy and other inherited genetic diseases.
Viral Vectors
Viral vectors are common carrier dependent delivery approaches used for gene editing purposes. Among these, adeno, adeno-associated, and lentiviral vectors have been extensively employed for the efficient loading of genetic materials to the target cells in both preclinical and clinical studies. These vectors act as highly efficient in vitro delivery approaches of genome-editing tools for both clinical and research applications [186]. Further details on the viral-mediated CRISPR/Cas9 delivery into different types of stem cells are presented in the following sections, and a summary of these studies is provided in Table 3.
Adenoviral Vectors (AdVs)
The AdVs are double-stranded DNA viruses with icosahedral nucleocapsids that are able to infect dividing and non-dividing cells. Owing to their advantages, AdVs are considered promising gene transfer vectors with high transduction efficiency and expression level of the transgene in mammalian cells. Upon injection, the AdVs genome remains extrachromosomal without integration into the host genome [205]. This is especially important for CRISPR/Cas9-mediated genome editing as it minimizes the possibility of insertional mutagenesis and off-target effects. Recently, significant efforts have been made to optimize AdVs for gene delivery applications. In the first attempt, recombinant AdVs were generated by the deletion of the E1 viral gene [206]. Next, a second generation of AdVs with 8 kb packaging capacity was developed through the deletion of E2 and E4 viral genes with the aim of decreasing the chronic immune responses [207, 208]. In the third generation of AdVs, coined as helper-dependent (HDAd) or gutless (GLAd), all the adenoviral genes were deleted to provide a higher packaging capacity of up to 37 kb for loading larger cargoes. These AdVs only contain inverted terminal repeat repeats and packaging signal (ѱ) required for DNA replication and encapsidation, respectively. These unique features make the HDAds an ideal option for encapsulating the CRISPR/Cas9 system in a single vector providing the benefit of delivery procedure simplicity [209, 210]. CRISPR/Cas9 delivery by AdV vectors has already been utilized in drug discovery, disease modeling, and treatments [118]. In 2018, non-integrating chimeric HDAds (Ad5/35++) were developed from serotype 5 of species C and serotype 35 of species B AdVs, respectively. The generated vectors interacted with CD46, a membrane protein constantly expressed on human CD34+ cells, resulting in efficient transduction of HSCs followed by high expression of CRISPR/Cas9 plasmid. The HDAds (Ad5/35++) vector included a number of mutations in the Ad35 fiber knob enhancing CD46 targeting (>25-fold), leading to highly efficient transduction at lower numbers of the multiplicity of infection for viral particles. These chimeric HDAdVs were used to reactivate the fetal γ-globin gene in sickle cell disease and β-thalassemia, affecting the viability, in vitro expansion, and differentiation of human CD34+ cells. To control the CRISPR/Cas9 activity, Li and colleagues generated chimeric vectors encapsulating anti-CRISPR (Acr) AcrII4 and AcrII2 peptides to target CRISPR/Cas9 complex (HDAd-Acr). The CD34+ cells that were consecutively loaded with HDAd-CRISPR and HDAd-Acr demonstrated a significantly higher engraftment rate. 10 weeks upon transplantation, the engrafted CD34+ cells had target site disruption frequencies similar to those of the pre-transplanted cells, demonstrating high viability and good survival of the genetically edited primitive HSCs [126]. Although AdV-mediated gene delivery does not initiate chronic immune responses, the viral capsid still has the chance of inducing acute phase immune responses. It is noteworthy that almost all humans have experienced the AdV infection since infancy, resulting in the production of neutralizing antibodies. Therefore immunogenicity is one of the challenges associated with using AdVs for gene therapy in a variety of human diseases [205, 211]. Different strategies have been proposed to overcome this challenge including manipulation of the vector genome with the aim of decreasing the immunogenicity and chemical protection to reduce the undesirable surface interactions [212].
Adeno-Associated Viral Vectors (AAVs)
AAVs, also known as non-enveloped viruses, are among the most popular viral vectors for CRISPR/Cas9-based gene editing purposes. Owing to the unique characteristics of AAVs, such as their good safety profile and therapeutic potential, they have been extensively utilized in gene therapy clinical trials [213]. Other advantages include mild immunogenicity and cytotoxicity observed at high doses of AAVs injection in animal models [213, 214]. Moreover, upon transduction, the AAVs genome usually remains episomal or extrachromosomal, which further integrates into hotspots of mitochondrial DNA and specific locus of the host genome at chromosome 19q13.4 called AAV integration site 1 (AAVS1) [215, 216]. These integrating sites are known as safe harbors without any contribution to tumorigenesis.
Furthermore, AAVs concameters have shown the ability to provide steady transgene expressions due to their long-lasting existence in non-dividing cells. Various AAV serotypes have been shown to be suitable for tissue-targeted gene delivery as well as CRISPR/Cas9-based genome editing in specific tissues. To this aim, AAV6 and AAV9 serotypes have been used for genome editing in murine muscle and brain tissue, respectively [118]. In a study, Xu et al. leveraged the site-specific integration of AAVs to induce long-lasting expression of human blood-coagulating factor IX in transgenic mice [217]. In a recent study, Martin and colleagues proposed a new marker-free approach for genome editing of human pluripotent stem cells (hPSCs) through electroporation of CRISPR/Cas9 RNPs followed by AAV6-mediated donor template delivery. Using this approach, they achieved up to 94% mono-allelic edition frequency of sickle cell mutation at the hemoglobin beta (HBB) site in the hPSCs without requiring marker selection [195]. Despite the popularity of AAVs in gene editing clinical trials, the major drawback attributed to these vectors is their limited cloning capacity. Further investigations can focus on the generation of recombinant AAVs with a higher packaging capacity of transgene than the wild type to attenuate the impact of AAVs in clinical gene therapy.
Lentiviral Vectors (LVs)
LVs are considered highly efficient vectors for CRISPR/Cas9 delivery into a wide range of dividing and non-dividing cells for gene therapy of monogenic disorders. These vectors can provide the following advantages: i: higher capacity for packaging the transgene, ii) efficient transduction of a different type of dividing and non-dividing cells, iii) low immunogenicity and cytotoxicity, and iv) minimal effect on the cells cycle. These favorable aspects have made LVs the vector of choice for gene-editing of infections associated with hepatitis B virus (HBV), human immunodeficiency virus (HIV-1), and herpes simplex virus (HSV-1), as well as correcting the defects in human genetic disorders like cystic fibrosis [218,219,220]. Despite these appealing features, LVs suffer from issues such as off-target effects. Since LV-mediated delivery can result in stable expression of the CRISPR/Cas9, it may increase the chance of non-specific DSBs, unwanted off-target effects, and higher indels at off-target sites hindering their application for precise genome editing purposes [220]. To address these issues, integrase-deficient LVs (IDLVs) have been developed for efficient CRISPR/Cas9 delivery with superior cloning capacity while demonstrating a very weak integration capability and transient expression in the host cell [220]. In 2021, an IDLV-mediated CRISPR/Cas9 gene editing approach, coined as an “all-in-one” delivery system, was developed to encode guide RNA and donor DNA templates. Using this strategy, one-time correction of sickle cell disease mutation in the HBB gene was successfully achieved with an efficiency of up to 42% [199]. The application of the IDLVs for CRISPR/Cas9 delivery into the HSCs is yet to be extensively explored.
Virus-Like Particles (VLPs)
Although viral vectors have shown success in the efficient delivery of the CRISPR/Cas9 system into the desired cells, major weaknesses attributed to these vectors are the i) high expression level of Cas9 nuclease, ii) increased chance of off-target effects, and iii) immune and inflammatory response issues, and iv) integration into the host genome. Recently, a new type of delivery particle, virus-like particles (VLPs), has been developed for gene editing purposes [221]. These particles are derived from viruses and mimic them in size and shape. The VLPs contain almost all the viral components (e.g., capsid and envelope) except the genome, which eliminates the risk of genome integration and infection in the host cells [222]. These particles are mostly derived from LVs and can package different payloads, including mRNAs, proteins, and RNPs. VLPs take advantage of the high infecting efficiency of viral vectors for transient expression of the Cas9 nuclease resulting in a safe and highly efficient genome editing procedure [221, 222].
Recently, a new type of vesicle, called VEsiCas, was developed to deliver CRISPR/Cas9 RNPs into iPSCs effectively. These VLPs were derived from vesicular stomatitis viruses (VSV) and decorated with fusogenic glycoprotein (VSV-G) for efficient protein delivery. Since VSV-G-enveloped SpCas9 vesicles are free from viral DNA encoding sgRNA and SpCas9, it makes it possible to rapidly clear the nuclease components of the target cells that correlate with reduced genome-wide off-target cleavages. It is shown that in comparison with the classical method of SpCas9 RNPs electroporation for obtaining the transient SpCas9 activities, VEsiCas have lower toxic effects and higher efficiency in nuclease delivery [159].
A follow-up study has reported on the development of engineered murine leukemia VLPs loaded with Cas9-sgRNA RNPs (Nanoblades) for effective genome-editing in primary cells and cell lines such as human hematopoietic and human induced pluripotent stem cells [157]. Moreover, in vivo genome-editing was achieved by transgene-free Nanoblades in the liver of the injected mice and the mouse embryos. The complex of Nanoblades with donor DNA can also be used for homology-directed repair or may be programmed with modified variants of Cas9 for mediating transcriptional gene up-regulations. These engineered VLPs can be easily prepared and are affordable and easy-to-implement in cellular biology laboratories. Since Cas9-sgRNA complex delivery with these VLPs is transient and dose-dependent, it can reduce off-target effects compared with the commonly used CRISPR plasmid transfection method [157].
Challenges and Future Directions
Despite the advantages of exploiting the CRISPR/Cas9 system for gene editing purposes, extensive research is still required to determine the safety and editing efficiency achieved through this system. The main drawback associated with CRISPR/Cas9-mediated gene editing is the cleavage of off-target genomic sites, as a shorter sequence of the target RNA is exploited in this approach compared to those used in ZFN- and TALEN-mediated gene editing [223]. Although different strategies have been employed to increase the specificity of CRISPR/Cas9-mediated gene editing, such as advancements in gRNA design [224], the development of new versions of the Cas9 nuclease [225], and optimization of the CRISPR/Cas9 delivery mechanisms, off-target effects remain an important hinder to the clinical translation of CRISPR/Cas9-mediated gene editing. In addition, unintentional large deletions and complex genomic rearrangements have been observed in the CRISPR/Cas9 edited cells [226]. On the other hand, inherent individual human genetic variations (e.g., single nucleotide polymorphisms and copy number variations) can lead to unintentional off-target gene editing. Although the standard human genome is often used as a reference for CRISPR/Cas9 optimization and off-target tool design, these polymorphisms can lead to multiple off-targets even using well-designed gRNAs [227]. Therefore, genome-wide sequence analysis, large-scale off-target prediction, individual intensive genotoxicity risk assessment, and careful patient monitoring are the measures to be considered in CRISPR/Cas9-mediated gene editing. The immunogenicity of the Cas9 nuclease also needs to be considered during the clinical translation of the CRISPR/Cas9 technology, which can cause severe immune responses in patients treated with CRISPR/Cas9 edited stem cells [228].
Base-Editing and Prime-Editing as Novel Gene Editing Approaches
The CRISPR/Cas9-mediated genome editing relies on the induction of a DNA double-strand break in the target DNA sequence. The generation of these breaks raises several concerns about the clinical applications of this gene editing strategy. DNA base-editing and prime-editing are novel gene editing approaches proposed to overcome the field’s current challenges. DNA base-editor is an engineered Cas enzyme that can bind to the target site and modify the target nucleotide without generating DSBs. Adenine base-editor (ABE) and cytosine base-editor (CBE) are two well-known types of DNA base-editor systems. Until recently, the correction of four transition mutations (C → T, G → A, A → G, and T → C) was only possible using the known CRISPR/Cas9 base editing. A recent study has proposed two new base editors for efficient C to G transversion [229]. In addition, dual base-editor Cas enzymes have been engineered recently for combinatorial editing in human cells [73, 230, 231]. Taken together, the engineered base editors broaden the application of DNA base-editing to transversion mutations and more complex edits that are impossible using single DNA base-editors.
Prime-editing is another gene editing strategy recently developed to expand the range of mutations that can be edited [232]. Unlike CRISPR-based editing, prime-editing does not require double-strand DNA breaks. The prime-editing system consists of a Cas9 H840A nickase fused to an engineered reverse transcriptase enzyme and a prime editing gRNA (pegRNA). The pegRNA is an extended guide RNA that includes a primer binding site (PBS) and a reverse transcriptase (RT) template sequence, which is then reverse-transcribed to DNA by the RT enzyme. Prime editing has several benefits over CRISPR/Cas9-mediated gene editing, including the ability to introduce nearly all conceivable nucleotide substitutions, the absence of a need for simultaneous delivery of a corrective donor template, the elimination of indel-induced frameshifts, and a lower rate of off-target edits. Consequently, it is a new and ideal candidate for overcoming the limitations of current CRISPR/Cas9- mediated gene editing, which can open up promising avenues toward more versatile and improved genome editing [233].
Conclusion
Different factors can affect the efficiency of CRISPR/Cas9-mediated gene editing, and the delivery approach is an essential factor that plays a vital role in the efficient gene editing process, especially in the case of hard-to-transfect stem cells. Since gene editing in stem cells can translate to the clinic, developing effective methods for delivering gene-editing components to different types of stem cells is necessary. This review paper entails a wide range of physical, chemical, mechanical, viral, and nanoparticle-based methods used to deliver gene editing tools to stem cells. These methods categorize into two main classes, carrier-dependent and carrier independent delivery approaches. Amongst them, a subset of carrier independent delivery approaches, called physical delivery (e.g., electroporation), is the most widely used method that gained much more attention for delivery applications. Several efforts have been made to enhance the efficiency of loading gene editing tools into target cells by combining some of the aforementioned methods. Since gene editing approaches are evolving rapidly, choosing an effective mechanism for loading the newly developed gene editing tools into the target cells among the existing options relies on the aims and objectives of the experiments. Developing novel delivery technologies that efficiently deliver gene editing tools inside the target cells without affecting the cell functionality will significantly impact the research and clinical outcomes of stem cell gene editing with remarkable accuracy.
Data Availability
This is a narrative review based on published data.
Abbreviations
- CRISPR/Cas9 :
-
clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9
- hPSCs :
-
human pluripotent stem cells, indel: insertion or deletion
- DSBs :
-
double-strand breaks
- NHEJ :
-
non-homologous end-joining
- HDR :
-
homology-directed repair
- TALENs :
-
transcription activator-like effector nucleases
- ZFNs :
-
zinc-finger nucleases
- sgRNA :
-
single guide RNA
- crRNA :
-
CRISPR RNA
- tracrRNA :
-
trans-activating crRNA
- RNP :
-
ribonucleoprotein
- iTOP :
-
induced transduction by osmocytosis and propanebetaine
- iPSCs :
-
induced pluripotent stem cells
- DDEB :
-
dominant dystrophic epidermolysis bullosa
- RDEB :
-
recessive dystrophic epidermolysis bullosa
- ssODN :
-
single-stranded oligodeoxynucleotides
- ABE :
-
adenine based editor
- PSCs :
-
pluripotent stem cells
- HSCs :
-
hematopoietic stem cells
- CBE :
-
cytosine base editor
- NB-Chip :
-
nano-blade chip
- TRIAMF :
-
transmembrane internalization assisted by membrane filtration
- NPs :
-
nanoparticles
- LNPs :
-
lipid-based NPs
- FDA :
-
food and drug administration
- BrS :
-
Brugada syndrome
- PLGA :
-
poly lactic-co-glycolic acid
- EMA :
-
European medicine agency
- HSPCs :
-
hematopoietic stem and progenitor cells
- EVs :
-
extracellular vesicles
- AdVs :
-
adenoviral vectors
- HDAd :
-
helper-dependent adenovirus
- GLAd :
-
gutless adenovirus
- Acr :
-
anti-CRISPR
- AAVs :
-
adeno-associated viruses
- AAVS1 :
-
AAV integration site 1
- HBB :
-
hemoglobin beta
- LVs :
-
lentiviruses
- HBV :
-
hepatitis B virus
- HIV-1 :
-
human immunodeficiency virus-1
- HSV-1 :
-
herpes simplex virus
- IDLVs :
-
integrase-deficient LVs
- CRISPRa :
-
CRISPR activator
- CRISPR/dCas9-E :
-
CRISPR/dcas9-effector
- VLPs :
-
virus-like particles
- VSVs :
-
vesicular stomatitis viruses
- VSV-G :
-
vesicular stomatitis viruses decorated with fusogenic glycoprotein
- pegRNA :
-
prime editing gRNA
- gRNA :
-
guide RNA
- PBS :
-
primer binding site
- RT :
-
reverse transcriptase
References
Guha, T. K., Wai, A., & Hausner, G. (2017). Programmable genome editing tools and their regulation for efficient genome engineering. Computational and Structural Biotechnology Journal, 15, 146–160.
Carroll, D. (2014). Genome engineering with targetable nucleases. Annual Review of Biochemistry, 83(1), 409–439.
Lotfi, M., Ashouri, A., Mojarrad, M., Mozaffari-Jovin, S., & Abbaszadegan, M. R. (2023). Design principles of a novel construct for HBB gene-editing and investigation of its gene-targeting efficiency in HEK293 cells. Molecular Biotechnology, 1–14.
Li, H., Yang, Y., Hong, W., Huang, M., Wu, M., & Zhao, X. (2020). Applications of genome editing technology in the targeted therapy of human diseases: Mechanisms, advances and prospects. Signal Transduction and Targeted Therapy, 5(1), 1–23.
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science, 339(6121), 819–823.
Schwank, G., Koo, B.-K., Sasselli, V., Dekkers, J. F., Heo, I., Demircan, T., et al. (2013). Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell, 13(6), 653–658.
Firth, A. L., Menon, T., Parker, G. S., Qualls, S. J., Lewis, B. M., Ke, E., et al. (2015). Functional gene correction for cystic fibrosis in lung epithelial cells generated from patient iPSCs. Cell Reports, 12(9), 1385–1390.
Dever, D. P., Bak, R. O., Reinisch, A., Camarena, J., Washington, G., Nicolas, C. E., et al. (2016). CRISPR/Cas9 β-globin gene targeting in human haematopoietic stem cells. Nature, 539(7629), 384–389.
Frangoul, H., Altshuler, D., Cappellini, M. D., Chen, Y.-S., Domm, J., Eustace, B. K., et al. (2021). CRISPR-Cas9 gene editing for sickle cell disease and β-thalassemia. New England Journal of Medicine, 384(3), 252–260.
Shin, J. W., Kim, K.-H., Chao, M. J., Atwal, R. S., Gillis, T., MacDonald, M. E., et al. (2016). Permanent inactivation of Huntington's disease mutation by personalized allele-specific CRISPR/Cas9. Human Molecular Genetics, 25(20), 4566–4576.
Monteys, A. M., Ebanks, S. A., Keiser, M. S., & Davidson, B. L. (2017). CRISPR/Cas9 editing of the mutant huntingtin allele in vitro and in vivo. Molecular Therapy, 25(1), 12–23.
Ousterout, D. G., Kabadi, A. M., Thakore, P. I., Majoros, W. H., Reddy, T. E., & Gersbach, C. A. (2015). Multiplex CRISPR/Cas9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nature Communications, 6(1), 1–13.
Li, H. L., Fujimoto, N., Sasakawa, N., Shirai, S., Ohkame, T., Sakuma, T., et al. (2015). Precise correction of the dystrophin gene in duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Reports, 4(1), 143–154.
Salmaninejad, A., Jafari Abarghan, Y., Bozorg Qomi, S., Bayat, H., Yousefi, M., Azhdari, S., et al. (2021). Common therapeutic advances for Duchenne muscular dystrophy (DMD). International Journal of Neuroscience, 131(4), 370–389.
Flynn, R., Grundmann, A., Renz, P., Hänseler, W., James, W. S., Cowley, S. A., et al. (2015). CRISPR-mediated genotypic and phenotypic correction of a chronic granulomatous disease mutation in human iPS cells. Experimental Hematology, 43(10), 838–48. e3.
De Ravin, S. S., Li, L., Wu, X., Choi, U., Allen, C., Koontz, S., et al. (2017). CRISPR-Cas9 gene repair of hematopoietic stem cells from patients with X-linked chronic granulomatous disease. Science Translational Medicine, 9(372), eaah3480.
Buskin, A., Zhu, L., Chichagova, V., Basu, B., Mozaffari-Jovin, S., Dolan, D., et al. (2018). Disrupted alternative splicing for genes implicated in splicing and ciliogenesis causes PRPF31 retinitis pigmentosa. Nature Communications, 9(1), 1–19.
Park, C.-Y., Kim, D. H., Son, J. S., Sung, J. J., Lee, J., Bae, S., et al. (2015). Functional correction of large factor VIII gene chromosomal inversions in hemophilia a patient-derived iPSCs using CRISPR-Cas9. Cell Stem Cell, 17(2), 213–220.
Guan, Y., Ma, Y., Li, Q., Sun, Z., Ma, L., Wu, L., et al. (2016). CRISPR/Cas9-mediated somatic correction of a novel coagulator factor IX gene mutation ameliorates hemophilia in mouse. EMBO Molecular Medicine, 8(5), 477–488.
Patsali, P., Mussolino, C., Ladas, P., Floga, A., Kolnagou, A., Christou, S., et al. (2019). The scope for thalassemia gene therapy by disruption of aberrant regulatory elements. Journal of Clinical Medicine, 8(11), 1959.
Shariati, L., Rohani, F., Heidari Hafshejani, N., Kouhpayeh, S., Boshtam, M., Mirian, M., et al. (2018). Disruption of SOX6 gene using CRISPR/Cas9 technology for gamma-globin reactivation: An approach towards gene therapy of β-thalassemia. Journal of Cellular Biochemistry, 119(11), 9357–9363.
Khosravi, M. A., Abbasalipour, M., Concordet, J.-P., Vom Berg, J., Zeinali, S., Arashkia, A., et al. (2019). Targeted deletion of BCL11A gene by CRISPR-Cas9 system for fetal hemoglobin reactivation: A promising approach for gene therapy of beta thalassemia disease. European Journal of Pharmacology, 854, 398–405.
Sun, W., Ji, W., Hall, J. M., Hu, Q., Wang, C., Beisel, C. L., et al. (2015). Self-assembled DNA nanoclews for the efficient delivery of CRISPR–Cas9 for genome editing. Angewandte Chemie, 127(41), 12197–12201.
Liu, C., Zhang, L., Liu, H., & Cheng, K. (2017). Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications. Journal of Controlled Release, 266, 17–26.
Hecker, J. G. (2016). Non-viral, lipid-mediated DNA and mRNA gene therapy of the central nervous system (CNS): Chemical-based transfection. Gene Therapy for Neurological Disorders (pp. 307–324). Springer.
Lino, C. A., Harper, J. C., Carney, J. P., & Timlin, J. A. (2018). Delivering CRISPR: A review of the challenges and approaches. Drug Delivery, 25(1), 1234–1257.
Lin, Y.-X., Wang, Y., Blake, S., Yu, M., Mei, L., Wang, H., et al. (2020). RNA Nanotechnology-Mediated Cancer Immunotherapy Theranostics, 10(1), 281.
Kouranova, E., Forbes, K., Zhao, G., Warren, J., Bartels, A., Wu, Y., et al. (2016). CRISPRs for optimal targeting: Delivery of CRISPR components as DNA, RNA, and protein into cultured cells and single-cell embryos. Human Gene Therapy, 27(6), 464–475.
Zhang, Z., Zhang, Y., Gao, F., Han, S., Cheah, K. S., Tse, H.-F., et al. (2017). CRISPR/Cas9 genome-editing system in human stem cells: Current status and future prospects. Molecular Therapy-Nucleic Acids, 9, 230–241.
Matano, M., Date, S., Shimokawa, M., Takano, A., Fujii, M., Ohta, Y., et al. (2015). Modeling colorectal cancer using CRISPR-Cas9–mediated engineering of human intestinal organoids. Nature Medicine, 21(3), 256–262.
Fessler, E., Drost, J., van Hooff, S. R., Linnekamp, J. F., Wang, X., Jansen, M., et al. (2016). TGFβ signaling directs serrated adenomas to the mesenchymal colorectal cancer subtype. EMBO Molecular Medicine, 8(7), 745–760.
Morshedi Rad, D., Alsadat Rad, M., Razavi Bazaz, S., Kashaninejad, N., Jin, D., & Ebrahimi, W. M. (2021). A comprehensive review on intracellular delivery. Advanced Materials, 33(13), 2005363.
Stewart, M. P., Sharei, A., Ding, X., Sahay, G., Langer, R., & Jensen, K. F. (2016). In vitro and ex vivo strategies for intracellular delivery. Nature, 538(7624), 183–192.
Kim, S., Kim, D., Cho, S. W., Kim, J., & Kim, J.-S. (2014). Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Research, 24(6), 1012–1019.
Kholosy, W. M., Visscher, M., Ogink, K., Buttstedt, H., Griffin, K., Beier, A., et al. (2021). Simple, fast and efficient iTOP-mediated delivery of CRISPR/Cas9 RNP in difficult-to-transduce human cells including primary T cells. Journal of Biotechnology, 338, 71–80.
Peyravian, N., Malekzadeh Kebria, M., Kiani, J., Brouki Milan, P., & Mozafari, M. (2021). CRISPR-associated (CAS) effectors delivery via microfluidic cell-deformation Chip. Materials, 14(12), 3164.
Wells, D. (2004). Gene therapy progress and prospects: Electroporation and other physical methods. Gene Therapy, 11(18), 1363–1369.
Meacham, J. M., Durvasula, K., Degertekin, F. L., & Fedorov, A. G. (2014). Physical methods for intracellular delivery: Practical aspects from laboratory use to industrial-scale processing. Journal of Laboratory Automation, 19(1), 1–18.
Shi, J., Ma, Y., Zhu, J., Chen, Y., Sun, Y., Yao, Y., et al. (2018). A review on electroporation-based intracellular delivery. Molecules., 23(11), 3044.
Young, J. L., & Dean, D. A. (2015). Electroporation-mediated gene delivery. Advances in Genetics, 89, 49–88.
Kim, J. A., Cho, K., Shin, M. S., Lee, W. G., Jung, N., Chung, C., et al. (2008). A novel electroporation method using a capillary and wire-type electrode. Biosensors & Bioelectronics, 23(9), 1353–1360.
Supharattanasitthi, W., Carlsson, E., Sharif, U., & Paraoan, L. (2019). CRISPR/Cas9-mediated one step bi-allelic change of genomic DNA in iPSCs and human RPE cells in vitro with dual antibiotic selection. Scientific Reports, 9(1), 174.
Xu, X., Gao, D., Wang, P., Chen, J., Ruan, J., Xu, J., et al. (2018). Efficient homology-directed gene editing by CRISPR/Cas9 in human stem and primary cells using tube electroporation. Scientific Reports, 8(1), 1–11.
Paquet, D., Kwart, D., Chen, A., Sproul, A., Jacob, S., Teo, S., et al. (2016). Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature, 533(7601), 125–129.
Yudovich, D., Bäckström, A., Schmiderer, L., Žemaitis, K., Subramaniam, A., & Larsson, J. (2020). Combined lentiviral-and RNA-mediated CRISPR/Cas9 delivery for efficient and traceable gene editing in human hematopoietic stem and progenitor cells. Scientific Reports, 10(1), 1–11.
Lakshmipathy, U., Pelacho, B., Sudo, K., Linehan, J. L., Coucouvanis, E., Kaufman, D. S., et al. (2004). Efficient transfection of embryonic and adult stem cells. Stem Cells, 22(4), 531–543.
Levetzow, G. V., Spanholtz, J., Beckmann, J., Fischer, J., Kögler, G., Wernet, P., et al. (2006). Nucleofection, an efficient nonviral method to transfer genes into human hematopoietic stem and progenitor cells. Stem Cells and Development, 15(2), 278–285.
Han, S. Y., Gai, W., Yancovitz, M., Osman, I., Di Como, C. J., & Polsky, D. (2008). Nucleofection is a highly effective gene transfer technique for human melanoma cell lines. Experimental Dermatology, 17(5), 405–411.
Shinkuma, S., Guo, Z., & Christiano, A. M. (2016). Site-specific genome editing for correction of induced pluripotent stem cells derived from dominant dystrophic epidermolysis bullosa. Proceedings of the National Academy of Sciences, 113(20), 5676–5681.
Jacków, J., Guo, Z., Hansen, C., Abaci, H. E., Doucet, Y. S., Shin, J. U., et al. (2019). CRISPR/Cas9-based targeted genome editing for correction of recessive dystrophic epidermolysis bullosa using iPS cells. Proceedings of the National Academy of Sciences, 116(52), 26846–26852.
Ruan, J., Hirai, H., Yang, D., Ma, L., Hou, X., Jiang, H., et al. (2019). Efficient gene editing at major CFTR mutation loci. Molecular Therapy-Nucleic Acids, 16, 73–81.
Xu, H., Kita, Y., Bang, U., Gee, P., & Hotta, A. (2021). Optimized electroporation of CRISPR-Cas9/gRNA ribonucleoprotein complex for selection-free homologous recombination in human pluripotent stem cells. STAR Protocols, 2(4), 100965.
Kagita, A., Lung, M. S., Xu, H., Kita, Y., Sasakawa, N., Iguchi, T., et al. (2021). Efficient ssODN-mediated targeting by avoiding cellular inhibitory RNAs through precomplexed CRISPR-Cas9/sgRNA ribonucleoprotein. Stem Cell Reports, 16(4), 985–996.
Forbes, T. A., Howden, S. E., Lawlor, K., Phipson, B., Maksimovic, J., Hale, L., et al. (2018). Patient-iPSC-derived kidney organoids show functional validation of a ciliopathic renal phenotype and reveal underlying pathogenetic mechanisms. The American Journal of Human Genetics, 102(5), 816–831.
Bassuk, A. G., Zheng, A., Li, Y., Tsang, S. H., & Mahajan, V. B. (2016). Precision medicine: Genetic repair of retinitis pigmentosa in patient-derived stem cells. Scientific Reports, 6(1), 1–6.
Wang, L., Yi, F., Fu, L., Yang, J., Wang, S., Wang, Z., et al. (2017). CRISPR/Cas9-mediated targeted gene correction in amyotrophic lateral sclerosis patient iPSCs. Protein & Cell, 8(5), 365–378.
Wang, S., Min, Z., Ji, Q., Geng, L., Su, Y., Liu, Z., et al. (2020). Rescue of premature aging defects in Cockayne syndrome stem cells by CRISPR/Cas9-mediated gene correction. Protein & Cell, 11(1), 1–22.
Burnight, E. R., Gupta, M., Wiley, L. A., Anfinson, K. R., Tran, A., Triboulet, R., et al. (2017). Using CRISPR-Cas9 to generate gene-corrected autologous iPSCs for the treatment of inherited retinal degeneration. Molecular Therapy, 25(9), 1999–2013.
Kim, S.-I., Matsumoto, T., Kagawa, H., Nakamura, M., Hirohata, R., Ueno, A., et al. (2018). Microhomology-assisted scarless genome editing in human iPSCs. Nature Communications, 9(1), 1–14.
Xu, P., Tong, Y., Liu, X.-z., Wang, T.-t., Cheng, L., Wang, B.-y., et al. (2015). Both TALENs and CRISPR/Cas9 directly target the HBB IVS2–654 (C> T) mutation in β-thalassemia-derived iPSCs. Scientific Reports, 5(1), 1–12.
Chen, Y., Cao, J., Xiong, M., Petersen, A. J., Dong, Y., Tao, Y., et al. (2015). Engineering human stem cell lines with inducible gene knockout using CRISPR/Cas9. Cell Stem Cell, 17(2), 233–244.
Song, B., Fan, Y., He, W., Zhu, D., Niu, X., Wang, D., et al. (2015). Improved hematopoietic differentiation efficiency of gene-corrected beta-thalassemia induced pluripotent stem cells by CRISPR/Cas9 system. Stem Cells and Development, 24(9), 1053–1065.
Li, S., Zhang, A., Xue, H., Li, D., & Liu, Y. (2017). One-step piggyBac transposon-based CRISPR/Cas9 activation of multiple genes. Molecular Therapy-Nucleic Acids, 8, 64–76.
Chang, K.-H., Huang, C.-Y., Ou-Yang, C.-H., Ho, C.-H., Lin, H.-Y., Hsu, C.-L., et al. (2021). In vitro genome editing rescues parkinsonism phenotypes in induced pluripotent stem cells-derived dopaminergic neurons carrying LRRK2 p. G2019S mutation. Stem Cell Research & Therapy, 12(1), 1–18.
Howden, S. E., McColl, B., Glaser, A., Vadolas, J., Petrou, S., Little, M. H., et al. (2016). A Cas9 variant for efficient generation of indel-free knockin or gene-corrected human pluripotent stem cells. Stem Cell Reports, 7(3), 508–517.
Ding, Q., Regan, S. N., Xia, Y., Oostrom, L. A., Cowan, C. A., & Musunuru, K. (2013). Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell, 12(4), 393.
Wen, J., Tao, W., Hao, S., & Zu, Y. (2017). Cellular function reinstitution of offspring red blood cells cloned from the sickle cell disease patient blood post CRISPR genome editing. Journal of Hematology & Oncology, 10(1), 1–11.
Romero, Z., Lomova, A., Said, S., Miggelbrink, A., Kuo, C. Y., Campo-Fernandez, B., et al. (2019). Editing the sickle cell disease mutation in human hematopoietic stem cells: Comparison of endonucleases and homologous donor templates. Molecular Therapy, 27(8), 1389–1406.
Byambaa, S., Uosaki, H., Ohmori, T., Hara, H., Endo, H., Nureki, O., et al. (2021). Non-viral ex vivo genome-editing in mouse bona fide hematopoietic stem cells with CRISPR/Cas9. Molecular Therapy-Methods & Clinical Development, 20, 451–462.
Humbert, O., Radtke, S., Samuelson, C., Carrillo, R. R., Perez, A. M., Reddy, S. S., et al. (2019). Therapeutically relevant engraftment of a CRISPR-Cas9–edited HSC-enriched population with HbF reactivation in nonhuman primates. Science Translational Medicine, 11(503).
Newby, G. A., Yen, J. S., Woodard, K. J., Mayuranathan, T., Lazzarotto, C. R., Li, Y., et al. (2021). Base editing of haematopoietic stem cells rescues sickle cell disease in mice. Nature, 595(7866), 295–302.
Giani, F. C., Fiorini, C., Wakabayashi, A., Ludwig, L. S., Salem, R. M., Jobaliya, C. D., et al. (2016). Targeted application of human genetic variation can improve red blood cell production from stem cells. Cell Stem Cell, 18(1), 73–78.
Zhang, X., Zhu, B., Chen, L., Xie, L., Yu, W., Wang, Y., et al. (2020). Dual base editor catalyzes both cytosine and adenine base conversions in human cells. Nature Biotechnology, 38(7), 856–860.
Hong, S. G., Yada, R. C., Choi, K., Carpentier, A., Liang, T. J., Merling, R. K., et al. (2017). Rhesus iPSC safe harbor gene-editing platform for stable expression of transgenes in differentiated cells of all germ layers. Molecular Therapy, 25(1), 44–53.
Marthaler, A. G., Schmid, B., Tubsuwan, A., Poulsen, U. B., Engelbrecht, A. F., Mau-Holzmann, U. A., et al. (2016). Generation of an isogenic, gene-corrected control cell line of the spinocerebellar ataxia type 2 patient-derived iPSC line H196. Stem Cell Research, 16(1), 162–165.
Deng, W.-L., Gao, M.-L., Lei, X.-L., Lv, J.-N., Zhao, H., He, K.-W., et al. (2018). Gene correction reverses ciliopathy and photoreceptor loss in iPSC-derived retinal organoids from retinitis pigmentosa patients. Stem Cell Reports, 10(4), 1267–1281.
van der Wal, E., Herrero-Hernandez, P., Wan, R., Broeders, M., In't Groen, S. L., van Gestel, T. J., et al. (2018). Large-scale expansion of human iPSC-derived skeletal muscle cells for disease modeling and cell-based therapeutic strategies. Stem Cell Reports, 10(6), 1975–1990.
Wen, W., Cheng, X., Fu, Y., Meng, F., Zhang, J.-P., Zhang, L., et al. (2018). High-level precise knockin of iPSCs by simultaneous reprogramming and genome editing of human peripheral blood mononuclear cells. Stem Cell Reports, 10(6), 1821–1834.
Smith, C., Abalde-Atristain, L., He, C., Brodsky, B. R., Braunstein, E. M., Chaudhari, P., et al. (2015). Efficient and allele-specific genome editing of disease loci in human iPSCs. Molecular Therapy, 23(3), 570–577.
Huang, X., Wang, Y., Yan, W., Smith, C., Ye, Z., Wang, J., et al. (2015). Production of gene-corrected adult beta globin protein in human erythrocytes differentiated from patient iPSCs after genome editing of the sickle point mutation. Stem Cells, 33(5), 1470–1479.
Morishige, S., Mizuno, S., Ozawa, H., Nakamura, T., Mazahery, A., Nomura, K., et al. (2020). CRISPR/Cas9-mediated gene correction in hemophilia B patient-derived iPSCs. International Journal of Hematology, 111(2), 225–233.
Selvaraj, S., Dhoke, N. R., Kiley, J., Mateos-Aierdi, A. J., Tungtur, S., Mondragon-Gonzalez, R., et al. (2019). Gene correction of LGMD2A patient-specific iPSCs for the development of targeted autologous cell therapy. Molecular Therapy, 27(12), 2147–2157.
Liu, J., Gao, C., Chen, W., Ma, W., Li, X., Shi, Y., et al. (2016). CRISPR/Cas9 facilitates investigation of neural circuit disease using human iPSCs: Mechanism of epilepsy caused by an SCN1A loss-of-function mutation. Translational Psychiatry, 6(1), e703.
Yada, R. C., Ostrominski, J. W., Tunc, I., Hong, S. G., Zou, J., & Dunbar, C. E. (2017). CRISPR/Cas9-based safe-harbor gene editing in rhesus iPSCs. Current Protocols in Stem Cell Biology, 43(1), 5A. 11.1–5A. 11.4.
Cai, L., Bai, H., Mahairaki, V., Gao, Y., He, C., Wen, Y., et al. (2018). A universal approach to correct various HBB gene mutations in human stem cells for gene therapy of beta-thalassemia and sickle cell disease. Stem Cells Translational Medicine, 7(1), 87–97.
Grobarczyk, B., Franco, B., Hanon, K., & Malgrange, B. (2015). Generation of isogenic human iPS cell line precisely corrected by genome editing using the CRISPR/Cas9 system. Stem Cell Reviews and Reports, 11(5), 774–787.
Lee, P. C., Truong, B., Vega-Crespo, A., Gilmore, W. B., Hermann, K., Angarita, S. A., et al. (2016). Restoring ureagenesis in hepatocytes by CRISPR/Cas9-mediated genomic addition to arginase-deficient induced pluripotent stem cells. Molecular Therapy-Nucleic Acids, 5, e394.
Xie, F., Ye, L., Chang, J. C., Beyer, A. I., Wang, J., Muench, M. O., et al. (2014). Seamless gene correction of β-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac. Genome Research, 24(9), 1526–1533.
Lyu, C., Shen, J., Wang, R., Gu, H., Zhang, J., Xue, F., et al. (2018). Targeted genome engineering in human induced pluripotent stem cells from patients with hemophilia B using the CRISPR-Cas9 system. Stem Cell Research & Therapy, 9(1), 1–12.
Turan, S., Farruggio, A. P., Srifa, W., Day, J. W., & Calos, M. P. (2016). Precise correction of disease mutations in induced pluripotent stem cells derived from patients with limb girdle muscular dystrophy. Molecular Therapy, 24(4), 685–696.
Nimsanor, N., Kitiyanant, N., Poulsen, U., Rasmussen, M. A., Clausen, C., Mau-Holzmann, U. A., et al. (2016). Generation of an isogenic, gene-corrected iPSC line from a symptomatic 57-year-old female patient with frontotemporal dementia caused by a P301L mutation in the microtubule associated protein tau (MAPT) gene. Stem Cell Research, 17(3), 556–559.
van der Kant, R., Langness, V. F., Herrera, C. M., Williams, D. A., Fong, L. K., Leestemaker, Y., et al. (2019). Cholesterol metabolism is a druggable axis that independently regulates tau and amyloid-β in iPSC-derived Alzheimer’s disease neurons. Cell Stem Cell, 24(3), 363–75.e9.
Mitzelfelt, K. A., McDermott-Roe, C., Grzybowski, M. N., Marquez, M., Kuo, C.-T., Riedel, M., et al. (2017). Efficient precision genome editing in iPSCs via genetic co-targeting with selection. Stem Cell Reports, 8(3), 491–499.
Shuangyu M, Viola R, Sui L, Cherubini V, Barbetti F, Egli D. β Cell Replacement after Gene Editing of a Neonatal Diabetes-Causing Mutation at the Insulin Locus. 2018.
Klatt, D., Cheng, E., Philipp, F., Selich, A., Dahlke, J., Schmidt, R. E., et al. (2019). Targeted repair of p47-CGD in iPSCs by CRISPR/Cas9: Functional correction without cleavage in the highly homologous pseudogenes. Stem Cell Reports, 13(4), 590–598.
Qin, Y. (2019). Generation of non-integrated iPSCs from achondroplasia patient skin and urine and gene correction in them using CRISPR-Cas9. bioRxiv, 801415.
Chen, J., Tang, Z., Zheng, J., Shi, H., Ding, J., Qian, X., et al. (2016). Effects of genetic correction on the differentiation of hair cell-like cells from iPSCs with MYO15A mutation. Cell Death & Differentiation, 23(8), 1347–1357.
Wattanapanitch, M., Damkham, N., Potirat, P., Trakarnsanga, K., Janan, M., Kheolamai, P., et al. (2018). One-step genetic correction of hemoglobin E/beta-thalassemia patient-derived iPSCs by the CRISPR/Cas9 system. Stem Cell Research & Therapy, 9(1), 1–11.
Niu, X., He, W., Song, B., Ou, Z., Fan, D., Chen, Y., et al. (2016). Combining single strand oligodeoxynucleotides and CRISPR/Cas9 to correct gene mutations in β-thalassemia-induced pluripotent stem cells. Journal of Biological Chemistry, 291(32), 16576–16585.
Jiang, S., Wen, N., Li, Z., Dube, U., Del Aguila, J., Budde, J., et al. (2018). Integrative system biology analyses of CRISPR-edited iPSC-derived neurons and human brains reveal deficiencies of presynaptic signaling in FTLD and PSP. Translational Psychiatry, 8(1), 1–16.
Budde, J. P., Martinez, R., Hsu, S., Wen, N., Chen, J. A., Coppola, G., et al. (2017). Precision genome-editing with CRISPR/Cas9 in human induced pluripotent stem cells. bioRxiv, 187377.
Nimsanor, N., Poulsen, U., Rasmussen, M. A., Clausen, C., Mau-Holzmann, U. A., Nielsen, J. E., et al. (2016). Generation of an isogenic, gene-corrected iPSC line from a symptomatic 59-year-old female patient with frontotemporal dementia caused by an R406W mutation in the microtubule associated protein tau (MAPT) gene. Stem Cell Research, 17(3), 576–579.
Pires, C., Schmid, B., Petræus, C., Poon, A., Nimsanor, N., Nielsen, T. T., et al. (2016). Generation of a gene-corrected isogenic control cell line from an Alzheimer's disease patient iPSC line carrying a A79V mutation in PSEN1. Stem Cell Research, 17(2), 285–288.
Poon, A., Schmid, B., Pires, C., Nielsen, T. T., Hjermind, L. E., Nielsen, J. E., et al. (2016). Generation of a gene-corrected isogenic control hiPSC line derived from a familial Alzheimer's disease patient carrying a L150P mutation in presenilin 1. Stem Cell Research, 17(3), 466–469.
Ou, Z., Niu, X., He, W., Chen, Y., Song, B., Xian, Y., et al. (2016). The combination of CRISPR/Cas9 and iPSC technologies in the gene therapy of human β-thalassemia in mice. Scientific Reports, 6(1), 1–13.
He, Q., Wang, H., Cheng, T., Yuan, W., Ma, Y., Jiang, Y., et al. (2017). Genetic correction and hepatic differentiation of hemophilia B-specific human induced pluripotent stem cells. Chinese Medical Sciences Journal, 32(3), 135–144.
Frederiksen, H. R., Holst, B., Ramakrishna, S., Muddashetty, R., Schmid, B., & Freude, K. (2019). Generation of two iPSC lines with either a heterozygous V717I or a heterozygous KM670/671NL mutation in the APP gene. Stem Cell Research, 34, 101368.
Deneault, E., White, S. H., Rodrigues, D. C., Ross, P. J., Faheem, M., Zaslavsky, K., et al. (2018). Complete disruption of autism-susceptibility genes by gene editing predominantly reduces functional connectivity of isogenic human neurons. Stem Cell Reports, 11(5), 1211–1225.
Wu, M., Liu, S., Gao, Y., Bai, H., Machairaki, V., Li, G., et al. (2018). Conditional gene knockout and reconstitution in human iPSCs with an inducible Cas9 system. Stem Cell Research, 29, 6–14.
Mandegar, M. A., Huebsch, N., Frolov, E. B., Shin, E., Truong, A., Olvera, M. P., et al. (2016). CRISPR interference efficiently induces specific and reversible gene silencing in human iPSCs. Cell Stem Cell, 18(4), 541–553.
Rakovic, A., Domingo, A., Grütz, K., Kulikovskaja, L., Capetian, P., Cowley, S. A., et al. (2018). Genome editing in induced pluripotent stem cells rescues TAF1 levels in X-linked dystonia-parkinsonism. Movement Disorders, 33(7), 1108–1118.
Zhang, H., Shi, J., Hachet, M. A., Xue, C., Bauer, R. C., Jiang, H., et al. (2017). CRISPR/Cas9-mediated gene editing in human iPSC-derived macrophage reveals lysosomal acid lipase function in human macrophages—Brief report. Arteriosclerosis, Thrombosis, and Vascular Biology, 37(11), 2156–2160.
Long, C., Li, H., Tiburcy, M., Rodriguez-Caycedo, C., Kyrychenko, V., Zhou, H., et al. (2018). Correction of diverse muscular dystrophy mutations in human engineered heart muscle by single-site genome editing. Science Advances, 4(1), eaap9004.
Young, C. S., Hicks, M. R., Ermolova, N. V., Nakano, H., Jan, M., Younesi, S., et al. (2016). A single CRISPR-Cas9 deletion strategy that targets the majority of DMD patients restores dystrophin function in hiPSC-derived muscle cells. Cell Stem Cell, 18(4), 533–540.
Kyrychenko, V., Kyrychenko, S., Tiburcy, M., Shelton, J. M., Long, C., Schneider, J. W., et al. (2017). Functional correction of dystrophin actin binding domain mutations by genome editing. JCI Insight, 2(18).
Antoniani, C., Meneghini, V., Lattanzi, A., Felix, T., Romano, O., Magrin, E., et al. (2018). Induction of fetal hemoglobin synthesis by CRISPR/Cas9-mediated editing of the human β-globin locus. Blood, The Journal of the American Society of Hematology, 131(17), 1960–1973.
Xie, N., Gong, H., Suhl, J. A., Chopra, P., Wang, T., & Warren, S. T. (2016). Reactivation of FMR1 by CRISPR/Cas9-mediated deletion of the expanded CGG-repeat of the fragile X chromosome. PLoS One, 11(10), e0165499.
Xu, C. L., Ruan, M. Z. C., Mahajan, V. B., & Tsang, S. H. (2019). Viral Delivery Systems for CRISPR. Viruses, 11(1).
Sürün, D., Schneider, A., Mircetic, J., Neumann, K., Lansing, F., Paszkowski-Rogacz, M., et al. (2020). Efficient generation and correction of mutations in human iPS cells utilizing mRNAs of CRISPR base editors and prime editors. Genes, 11(5), 511.
Antony, J. S., Latifi, N., Haque, A., Lamsfus-Calle, A., Daniel-Moreno, A., Graeter, S., et al. (2018). Gene correction of HBB mutations in CD34(+) hematopoietic stem cells using Cas9 mRNA and ssODN donors. Molecular and Cellular Pediatrics, 5(1), 9.
Park, S. H., Lee, C. M., Dever, D. P., Davis, T. H., Camarena, J., Srifa, W., et al. (2019). Highly efficient editing of the β-globin gene in patient-derived hematopoietic stem and progenitor cells to treat sickle cell disease. Nucleic Acids Research, 47(15), 7955–7972.
Quintana-Bustamante, O., Fañanas-Baquero, S., Orman, I., Torres, R., Duchateau, P., Poirot, L., et al. (2019). Gene editing of PKLR gene in human hematopoietic progenitors through 5′ and 3' UTR modified TALEN mRNA. PLoS One, 14(10), e0223775.
DeWitt, M. A., Magis, W., Bray, N. L., Wang, T., Berman, J. R., Urbinati, F., et al. (2016). Selection-free genome editing of the sickle mutation in human adult hematopoietic stem/progenitor cells. Science Translational Medicine, 8(360), 360ra134.
Mettananda, S., Fisher, C. A., Hay, D., Badat, M., Quek, L., Clark, K., et al. (2017). Editing an α-globin enhancer in primary human hematopoietic stem cells as a treatment for β-thalassemia. Nature Communications, 8(1), 1–11.
Mandal, P. K., Ferreira, L. M., Collins, R., Meissner, T. B., Boutwell, C. L., Friesen, M., et al. (2014). Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Cell Stem Cell, 15(5), 643–652.
Li, C., Psatha, N., Gil, S., Wang, H., Papayannopoulou, T., & Lieber, A. (2018). HDAd5/35++ adenovirus vector expressing anti-CRISPR peptides decreases CRISPR/Cas9 toxicity in human hematopoietic stem cells. Molecular Therapy-Methods & Clinical Development, 9, 390–401.
Xu, H., Wang, B., Ono, M., Kagita, A., Fujii, K., Sasakawa, N., et al. (2019). Targeted disruption of HLA genes via CRISPR-Cas9 generates iPSCs with enhanced immune compatibility. Cell Stem Cell, 24(4), 566–78.e7.
Patsali, P., Turchiano, G., Papasavva, P., Romito, M., Loucari, C. C., Stephanou, C., et al. (2019). Correction of IVS I-110 (G> a) β-thalassemia by CRISPR/Cas-and TALEN-mediated disruption of aberrant regulatory elements in human hematopoietic stem and progenitor cells. Haematologica, 104(11), e497.
Wu, Y., Zeng, J., Roscoe, B. P., Liu, P., Yao, Q., Lazzarotto, C. R., et al. (2019). Highly efficient therapeutic gene editing of human hematopoietic stem cells. Nature Medicine, 25(5), 776–783.
Hendel, A., Bak, R. O., Clark, J. T., Kennedy, A. B., Ryan, D. E., Roy, S., et al. (2015). Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nature Biotechnology, 33(9), 985–989.
Wagenblast, E., Azkanaz, M., Smith, S. A., Shakib, L., McLeod, J. L., Krivdova, G., et al. (2019). Functional profiling of single CRISPR/Cas9-edited human long-term hematopoietic stem cells. Nature Communications, 10(1), 1–11.
Zeng, J., Wu, Y., Ren, C., Bonanno, J., Shen, A. H., Shea, D., et al. (2020). Therapeutic base editing of human hematopoietic stem cells. Nature Medicine, 26(4), 535–541.
Batista, P. J., Molinie, B., Wang, J., Qu, K., Zhang, J., Li, L., et al. (2014). m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell, 15(6), 707–719.
Li, Y., Rivera, C. M., Ishii, H., Jin, F., Selvaraj, S., Lee, A. Y., et al. (2014). CRISPR reveals a distal super-enhancer required for Sox2 expression in mouse embryonic stem cells. PLoS One, 9(12), e114485.
Hu, Y., Liu, S., & Zhu, B.-M. (2020). CRISPR/Cas9-induced loss of Keap1 enhances anti-oxidation in rat adipose-derived mesenchymal stem cells. Frontiers in Neurology, 1311.
Castaño, J., Bueno, C., Jiménez-Delgado, S., Roca-Ho, H., Fraga, M. F., Fernandez, A. F., et al. (2017). Generation and characterization of a human iPSC cell line expressing inducible Cas9 in the "safe harbor" AAVS1 locus. Stem Cell Research, 21, 137–140.
Hosseini Far, H., Patria, Y. N., Motazedian, A., Elefanty, A. G., Stanley, E. G., Lamandé, S. R., et al. (2019). Generation of a heterozygous COL1A1 (c.3969_3970insT) osteogenesis imperfecta mutation human iPSC line, MCRIi001-a-1, using CRISPR/Cas9 editing. Stem Cell Research, 37, 101449.
Tidball, A. M., Swaminathan, P., Dang, L. T., & Parent, J. M. (2018). Generating loss-of-function iPSC lines with combined CRISPR indel formation and reprogramming from human fibroblasts. Bio-Protocol, 8(7).
Bae, T., Hur, J. W., Kim, D., & Hur, J. K. (2019). Recent trends in CRISPR-Cas system: Genome, epigenome, and transcriptome editing and CRISPR delivery systems. Genes & Genomics, 41(8), 871–877.
Zuris, J. A., Thompson, D. B., Shu, Y., Guilinger, J. P., Bessen, J. L., Hu, J. H., et al. (2015). Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nature Biotechnology, 33(1), 73–80.
D'Astolfo, D. S., Pagliero, R. J., Pras, A., Karthaus, W. R., Clevers, H., Prasad, V., et al. (2015). Efficient intracellular delivery of native proteins. Cell, 161(3), 674–690.
Ramakrishna, S., Kwaku Dad, A. B., Beloor, J., Gopalappa, R., Lee, S. K., & Kim, H. (2014). Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Research, 24(6), 1020–1027.
Suresh, B., Ramakrishna, S., & Kim, H. (2017). Cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA for genome editing. Methods in Molecular Biology (Clifton, NJ), 1507, 81–94.
Morshedi Rad, D., Rezaei, M., Radfar, P., & Ebrahimi, W. M. (2022). Microengineered filters for efficient delivery of nanomaterials into mammalian cells. Scientific Reports, 12(1), 4383.
Ma, Y., Han, X., Quintana Bustamante, O., Bessa de Castro, R., Zhang, K., Zhang, P., et al. (2017). Highly efficient genome editing of human hematopoietic stem cells via a nano-silicon-blade delivery approach. Integrative Biology, 9(6), 548–554.
Yen, J., Fiorino, M., Liu, Y., Paula, S., Clarkson, S., Quinn, L., et al. (2018). TRIAMF: A new method for delivery of Cas9 ribonucleoprotein complex to human hematopoietic stem cells. Scientific Reports, 8(1), 1–11.
Yan, J., Kang, D. D., & Dong, Y. (2021). Harnessing lipid nanoparticles for efficient CRISPR delivery. Biomaterials Science.
Chen, F., Alphonse, M., & Liu, Q. (2020). Strategies for nonviral nanoparticle-based delivery of CRISPR/Cas9 therapeutics. Wiley Interdisciplinary Reviews: Nanomedicine and Nanobiotechnology, 12(3), e1609.
Pensado, A., Seijo, B., & Sanchez, A. (2014). Current strategies for DNA therapy based on lipid nanocarriers. Expert Opinion on Drug Delivery, 11(11), 1721–1731.
Yip, B. H. (2020). Recent advances in CRISPR/Cas9 delivery strategies. Biomolecules, 10(6), 839.
Allen, T. M., & Cullis, P. R. (2013). Liposomal drug delivery systems: From concept to clinical applications. Advanced Drug Delivery Reviews, 65(1), 36–48.
Liang, P., Sallam, K., Wu, H., Li, Y., Itzhaki, I., Garg, P., et al. (2016). Patient-specific and genome-edited induced pluripotent stem cell–derived cardiomyocytes elucidate single-cell phenotype of Brugada syndrome. Journal of the American College of Cardiology, 68(19), 2086–2096.
Yang, T.-C., Chang, C.-Y., Yarmishyn, A. A., Mao, Y.-S., Yang, Y.-P., Wang, M.-L., et al. (2020). Carboxylated nanodiamond-mediated CRISPR-Cas9 delivery of human retinoschisis mutation into human iPSCs and mouse retina. Acta Biomaterialia, 101, 484–494.
Gee, P., Lung, M. S., Okuzaki, Y., Sasakawa, N., Iguchi, T., Makita, Y., et al. (2020). Extracellular nanovesicles for packaging of CRISPR-Cas9 protein and sgRNA to induce therapeutic exon skipping. Nature Communications, 11(1), 1–18.
Lee, K., Conboy, M., Park, H. M., Jiang, F., Kim, H. J., Dewitt, M. A., et al. (2017). Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nature Biomedical Engineering, 1(11), 889–901.
Shahbazi, R., Sghia-Hughes, G., Reid, J. L., Kubek, S., Haworth, K. G., Humbert, O., et al. (2019). Targeted homology-directed repair in blood stem and progenitor cells with CRISPR nanoformulations. Nature Materials, 18(10), 1124–1132.
Mangeot, P. E., Risson, V., Fusil, F., Marnef, A., Laurent, E., Blin, J., et al. (2019). Genome editing in primary cells and in vivo using viral-derived Nanoblades loaded with Cas9-sgRNA ribonucleoproteins. Nature Communications, 10(1), 1–15.
Lin, Y., Wu, J., Gu, W., Huang, Y., Tong, Z., Huang, L., et al. (2018). Exosome–liposome hybrid nanoparticles deliver CRISPR/Cas9 system in MSCs. Advanced Science, 5(4), 1700611.
Montagna, C., Petris, G., Casini, A., Maule, G., Franceschini, G. M., Zanella, I., et al. (2018). VSV-G-enveloped vesicles for traceless delivery of CRISPR-Cas9. Molecular Therapy-Nucleic Acids, 12, 453–462.
Garg, P., Oikonomopoulos, A., Chen, H., Li, Y., Lam, C. K., Sallam, K., et al. (2018). Genome editing of induced pluripotent stem cells to decipher cardiac channelopathy variant. Journal of the American College of Cardiology, 72(1), 62–75.
Shao, Q., Esseltine, J. L., Huang, T., Novielli-Kuntz, N., Ching, J. E., Sampson, J., et al. (2019). Connexin43 is dispensable for early stage human mesenchymal stem cell adipogenic differentiation but is protective against cell senescence. Biomolecules, 9(9), 474.
Qi, T., Wu, F., Xie, Y., Gao, S., Li, M., Pu, J., et al. (2020). Base editing mediated generation of point mutations into human pluripotent stem cells for modeling disease. Frontiers in Cell and Developmental Biology, 1029.
Brunger, J. M., Zutshi, A., Willard, V. P., Gersbach, C. A., & Guilak, F. (2017). CRISPR/Cas9 editing of murine induced pluripotent stem cells for engineering inflammation-resistant tissues. Arthritis & Rheumatology, 69(5), 1111–1121.
Horii, T., Tamura, D., Morita, S., Kimura, M., & Hatada, I. (2013). Generation of an ICF syndrome model by efficient genome editing of human induced pluripotent stem cells using the CRISPR system. International Journal of Molecular Sciences, 14(10), 19774–19781.
Chen, Y., Spitzer, S., Agathou, S., Karadottir, R. T., & Smith, A. (2017). Gene editing in rat embryonic stem cells to produce in vitro models and in vivo reporters. Stem Cell Reports, 9(4), 1262–1274.
Shen, Y., Zhang, J., Yu, T., & Qi, C. (2018). Generation of PTEN knockout bone marrow mesenchymal stem cell lines by CRISPR/Cas9-mediated genome editing. Cytotechnology, 70(2), 783–791.
Wang, Y., Ding, Y., & Li, J. (2017). CRISPR-Cas9-mediated gene editing in mouse Spermatogonial stem cells. In B. Zhang (Ed.), RNAi and small regulatory RNAs in stem cells: Methods and protocols (pp. 293–305). Springer New York.
Meng, X., Zheng, M., Yu, M., Bai, W., Zuo, L., Bu, X., et al. (2019). Transplantation of CRISPRa system engineered IL10-overexpressing bone marrow-derived mesenchymal stem cells for the treatment of myocardial infarction in diabetic mice. Journal of Biological Engineering, 13(1), 1–12.
Hwang, S. J., Bellocq, N. C., & Davis, M. E. (2001). Effects of structure of beta-cyclodextrin-containing polymers on gene delivery. Bioconjugate Chemistry, 12(2), 280–290.
Felgner, P. L., Barenholz, Y., Behr, J. P., Cheng, S. H., Cullis, P., Huang, L., et al. (1997). Nomenclature for synthetic gene delivery systems. Human Gene Therapy, 8(5), 511–512.
Prokop, A., & Davidson, J. M. (2008). Nanovehicular intracellular delivery systems. Journal of Pharmaceutical Sciences, 97(9), 3518–3590.
Vert, M., Mauduit, J., & Li, S. (1994). Biodegradation of PLA/GA polymers: Increasing complexity. Biomaterials, 15(15), 1209–1213.
Wang, H. X., Song, Z., Lao, Y. H., Xu, X., Gong, J., Cheng, D., et al. (2018). Nonviral gene editing via CRISPR/Cas9 delivery by membrane-disruptive and endosomolytic helical polypeptide. Proceedings of the National Academy of Sciences of the United States of America, 115(19), 4903–4908.
Kumari, A., Yadav, S. K., & Yadav, S. C. (2010). Biodegradable polymeric nanoparticles based drug delivery systems. Colloids and Surfaces B, Biointerfaces, 75(1), 1–18.
Cruz, L. J., van Dijk, T., Vepris, O., Li, T., Schomann, T., Baldazzi, F., et al. (2021). PLGA-nanoparticles for intracellular delivery of the CRISPR-complex to elevate fetal globin expression in erythroid cells. Biomaterials, 268, 120580.
Loh, X. J., Lee, T. C., Dou, Q., & Deen, G. R. (2016). Utilising inorganic nanocarriers for gene delivery. Biomaterials Science, 4(1), 70–86.
Lee, B., Lee, K., Panda, S., Gonzales-Rojas, R., Chong, A., Bugay, V., et al. (2018). Nanoparticle delivery of CRISPR into the brain rescues a mouse model of fragile X syndrome from exaggerated repetitive behaviours. Nature Biomedical Engineering, 2(7), 497–507.
Zhang, X. (2015). Gold nanoparticles: Recent advances in the biomedical applications. Cell Biochemistry and Biophysics, 72(3), 771–775.
Mout, R., Ray, M., Yesilbag Tonga, G., Lee, Y. W., Tay, T., Sasaki, K., et al. (2017). Direct cytosolic delivery of CRISPR/Cas9-ribonucleoprotein for efficient gene editing. ACS Nano, 11(3), 2452–2458.
Lai, R. C., Yeo, R. W. Y., Tan, K. H., & Lim, S. K. (2013). Exosomes for drug delivery—A novel application for the mesenchymal stem cell. Biotechnology Advances, 31(5), 543–551.
Kao, C.-Y., & Papoutsakis, E. T. (2019). Extracellular vesicles: Exosomes, microparticles, their parts, and their targets to enable their biomanufacturing and clinical applications. Current Opinion in Biotechnology, 60, 89–98.
Choi, J. G., Dang, Y., Abraham, S., Ma, H., Zhang, J., Guo, H., et al. (2016). Lentivirus pre-packed with Cas9 protein for safer gene editing. Gene Therapy, 23(7), 627–633.
Campbell, L. A., Coke, L. M., Richie, C. T., Fortuno, L. V., Park, A. Y., & Harvey, B. K. (2019). Gesicle-mediated delivery of CRISPR/Cas9 ribonucleoprotein complex for inactivating the HIV provirus. Molecular Therapy, 27(1), 151–163.
Kooijmans, S. A., Vader, P., van Dommelen, S. M., van Solinge, W. W., & Schiffelers, R. M. (2012). Exosome mimetics: A novel class of drug delivery systems. International Journal of Nanomedicine, 7, 1525.
Ha, D., Yang, N., & Nadithe, V. (2016). Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: Current perspectives and future challenges. Acta Pharmaceutica Sinica B, 6(4), 287–296.
Yin, H., Kauffman, K. J., & Anderson, D. G. (2017). Delivery technologies for genome editing. Nature Reviews Drug Discovery, 16(6), 387–399.
Li, C., Psatha, N., Sova, P., Gil, S., Wang, H., Kim, J., et al. (2018). Reactivation of γ-globin in adult β-YAC mice after ex vivo and in vivo hematopoietic stem cell genome editing. Blood, The Journal of the American Society of Hematology, 131(26), 2915–2928.
Tabebordbar, M., Zhu, K., Cheng, J. K., Chew, W. L., Widrick, J. J., Yan, W. X., et al. (2016). In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science, 351(6271), 407–411.
Pavani, G., Laurent, M., Fabiano, A., Cantelli, E., Sakkal, A., Corre, G., et al. (2020). Ex vivo editing of human hematopoietic stem cells for erythroid expression of therapeutic proteins. Nature Communications, 11(1), 1–13.
Tran, N. T., Sommermann, T., Graf, R., Trombke, J., Pempe, J., Petsch, K., et al. (2019). Efficient CRISPR/Cas9-mediated gene knockin in mouse hematopoietic stem and progenitor cells. Cell Reports, 28(13), 3510–22.e5.
Rai, R., Romito, M., Rivers, E., Turchiano, G., Blattner, G., Vetharoy, W., et al. (2020). Targeted gene correction of human hematopoietic stem cells for the treatment of Wiskott-Aldrich syndrome. Nature Communications, 11(1), 1–15.
Gomez-Ospina, N., Scharenberg, S. G., Mostrel, N., Bak, R. O., Mantri, S., Quadros, R. M., et al. (2019). Human genome-edited hematopoietic stem cells phenotypically correct Mucopolysaccharidosis type I. Nature Communications, 10(1), 1–14.
Scharenberg, S. G., Poletto, E., Lucot, K. L., Colella, P., Sheikali, A., Montine, T. J., et al. (2020). Engineering monocyte/macrophage− specific glucocerebrosidase expression in human hematopoietic stem cells using genome editing. Nature Communications, 11(1), 1–14.
Lee, E., Goodwin, M., Lakshmanan, U., Shipp, S., Pavel-Dinu, M., Porteus, M., et al. (2020). Gene editing using CRISPR enables FOXP3 gene repair in HSPCs and IPEX patient T cells. Cytotherapy, 22(5), S20.
Martin, R. M., Ikeda, K., Cromer, M. K., Uchida, N., Nishimura, T., Romano, R., et al. (2019). Highly efficient and marker-free genome editing of human pluripotent stem cells by CRISPR-Cas9 RNP and AAV6 donor-mediated homologous recombination. Cell Stem Cell, 24(5), 821–8.e5.
Eggenschwiler, R., Moslem, M., Fráguas, M. S., Galla, M., Papp, O., Naujock, M., et al. (2016). Improved bi-allelic modification of a transcriptionally silent locus in patient-derived iPSC by Cas9 nickase. Scientific Reports, 6(1), 1–14.
Chang, C.-W., Lai, Y.-S., Westin, E., Khodadadi-Jamayran, A., Pawlik, K. M., Lamb, L. S., Jr., et al. (2015). Modeling human severe combined immunodeficiency and correction by CRISPR/Cas9-enhanced gene targeting. Cell Reports, 12(10), 1668–1677.
Ye, L., Wang, J., Tan, Y., Beyer, A. I., Xie, F., Muench, M. O., et al. (2016). Genome editing using CRISPR-Cas9 to create the HPFH genotype in HSPCs: An approach for treating sickle cell disease and β-thalassemia. Proceedings of the National Academy of Sciences, 113(38), 10661–10665.
Uchida, N., Drysdale, C. M., Nassehi, T., Gamer, J., Yapundich, M., DiNicola, J., et al. (2021). Cas9 protein delivery non-integrating lentiviral vectors for gene correction in sickle cell disease. Molecular Therapy-Methods & Clinical Development, 21, 121–132.
Hoban, M. D., Lumaquin, D., Kuo, C. Y., Romero, Z., Long, J., Ho, M., et al. (2016). CRISPR/Cas9-mediated correction of the sickle mutation in human CD34+ cells. Molecular Therapy, 24(9), 1561–1569.
Traxler, E. A., Yao, Y., Wang, Y.-D., Woodard, K. J., Kurita, R., Nakamura, Y., et al. (2016). A genome-editing strategy to treat β-hemoglobinopathies that recapitulates a mutation associated with a benign genetic condition. Nature Medicine, 22(9), 987–990.
Shalem, O., Sanjana, N. E., Hartenian, E., Shi, X., Scott, D. A., Mikkelsen, T. S., et al. (2014). Genome-scale CRISPR-Cas9 knockout screening in human cells. Science, 343(6166), 84–87.
Kearns, N. A., Genga, R. M., Enuameh, M. S., Garber, M., Wolfe, S. A., & Maehr, R. (2014). Cas9 effector-mediated regulation of transcription and differentiation in human pluripotent stem cells. Development, 141(1), 219–223.
Sun, S., Xiao, J., Huo, J., Geng, Z., Ma, K., Sun, X., et al. (2018). Targeting ectodysplasin promotor by CRISPR/dCas9-effector effectively induces the reprogramming of human bone marrow-derived mesenchymal stem cells into sweat gland-like cells. Stem Cell Research & Therapy, 9(1), 1–10.
Tatsis, N., & Ertl, H. C. (2004). Adenoviruses as vaccine vectors. Molecular Therapy, 10(4), 616–629.
Wilmott, R. W., Amin, R. S., Perez, C. R., Wert, S. E., Keller, G., Boivin, G. P., et al. (1996). Safety of adenovirus-mediated transfer of the human cystic fibrosis transmembrane conductance regulator cDNA to the lungs of nonhuman primates. Human Gene Therapy, 7(3), 301–318.
Armentano, D., Sookdeo, C. C., Hehir, K. M., Gregory, R. J., St George, J. A., Prince, G. A., et al. (1995). Characterization of an adenovirus gene transfer vector containing an E4 deletion. Human Gene Therapy, 6(10), 1343–1353.
Engelhardt, J. F., Ye, X., Doranz, B., & Wilson, J. M. (1994). Ablation of E2A in recombinant adenoviruses improves transgene persistence and decreases inflammatory response in mouse liver. Proceedings of the National Academy of Sciences of the United States of America, 91(13), 6196–6200.
Parks, R. J., Chen, L., Anton, M., Sankar, U., Rudnicki, M. A., & Graham, F. L. (1996). A helper-dependent adenovirus vector system: Removal of helper virus by Cre-mediated excision of the viral packaging signal. Proceedings of the National Academy of Sciences of the United States of America, 93(24), 13565–13570.
Palmer, D., & Ng, P. (2003). Improved system for helper-dependent adenoviral vector production. Molecular Therapy, 8(5), 846–852.
Rosewell, A., Vetrini, F., & Ng, P. (2011). Helper-Dependent Adenoviral Vectors. Journal of Genetic Syndromes & Gene Therapy, (Suppl 5).
Kreppel, F., & Hagedorn, C. (2021). Capsid and genome modification strategies to reduce the immunogenicity of adenoviral vectors. International Journal of Molecular Sciences, 22(5), 2417.
Lau, C. H., & Suh, Y. (2017). In vivo genome editing in animals using AAV-CRISPR system: Applications to translational research of human disease. F1000Research, 6, 2153.
Naso, M. F., Tomkowicz, B., Perry, W. L., 3rd, & Strohl, W. R. (2017). Adeno-associated virus (AAV) as a vector for gene therapy. BioDrugs, 31(4), 317–334.
Kaeppel, C., Beattie, S. G., Fronza, R., van Logtenstein, R., Salmon, F., Schmidt, S., et al. (2013). A largely random AAV integration profile after LPLD gene therapy. Nature Medicine, 19(7), 889–891.
Giraud, C., Winocour, E., & Berns, K. I. (1994). Site-specific integration by adeno-associated virus is directed by a cellular DNA sequence. Proceedings of the National Academy of Sciences of the United States of America, 91(21), 10039–10043.
Xu, Z. X., Chen, J. Z., Yue, Y. B., Zhang, J. Q., Li, Z. H., Feng, D. M., et al. (2009). A 16-bp RBE element mediated rep-dependent site-specific integration in AAVS1 transgenic mice for expression of hFIX. Gene Therapy, 16(5), 589–595.
Kantor, B., Bailey, R. M., Wimberly, K., Kalburgi, S. N., & Gray, S. J. (2014). Methods for gene transfer to the central nervous system. Advances in Genetics, 87, 125–197.
Milone, M. C., & O’Doherty, U. (2018). Clinical use of lentiviral vectors. Leukemia, 32(7), 1529–1541.
Ortinski, P. I., O’Donovan, B., Dong, X., & Kantor, B. (2017). Integrase-deficient Lentiviral vector as an all-in-one platform for highly efficient CRISPR/Cas9-mediated gene editing. Molecular Therapy - Methods & Clinical Development, 5, 153–164.
Lyu, P., Wang, L., & Lu, B. (2020). Virus-like particle mediated CRISPR/Cas9 delivery for efficient and safe genome editing. Life, 10(12), 366.
Nooraei, S., Bahrulolum, H., Hoseini, Z. S., Katalani, C., Hajizade, A., Easton, A. J., et al. (2021). Virus-like particles: Preparation, immunogenicity and their roles as nanovaccines and drug nanocarriers. Journal of Nanobiotechnology, 19(1), 1–27.
Zischewski, J., Fischer, R., & Bortesi, L. (2017). Detection of on-target and off-target mutations generated by CRISPR/Cas9 and other sequence-specific nucleases. Biotechnology Advances, 35(1), 95–104.
Fu, Y., Sander, J. D., Reyon, D., Cascio, V. M., & Joung, J. K. (2014). Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nature Biotechnology, 32(3), 279–284.
Huang, X., Yang, D., Zhang, J., Xu, J., & Chen, Y. E. (2022). Recent advances in improving gene-editing specificity through CRISPR–Cas9 nuclease engineering. Cells, 11(14), 2186.
Shin, H. Y., Wang, C., Lee, H. K., Yoo, K. H., Zeng, X., Kuhns, T., et al. (2017). CRISPR/Cas9 targeting events cause complex deletions and insertions at 17 sites in the mouse genome. Nature Communications, 8(1), 1–10.
Scott, D. A., & Zhang, F. (2017). Implications of human genetic variation in CRISPR-based therapeutic genome editing. Nature medicine., 23(9), 1095–1101.
Charlesworth, C. T., Deshpande, P. S., Dever, D. P., Camarena, J., Lemgart, V. T., Cromer, M. K., et al. (2019). Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nature Medicine, 25(2), 249–254.
Kurt, I. C., Zhou, R., Iyer, S., Garcia, S. P., Miller, B. R., Langner, L. M., et al. (2021). CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nature Biotechnology, 39(1), 41–46.
Grünewald, J., Zhou, R., Lareau, C. A., Garcia, S. P., Iyer, S., Miller, B. R., et al. (2020). A dual-deaminase CRISPR base editor enables concurrent adenine and cytosine editing. Nature Biotechnology, 38(7), 861–864.
Sakata, R. C., Ishiguro, S., Mori, H., Tanaka, M., Tatsuno, K., Ueda, H., et al. (2020). Base editors for simultaneous introduction of C-to-T and A-to-G mutations. Nature Biotechnology, 38(7), 865–869.
Anzalone, A. V., Randolph, P. B., Davis, J. R., Sousa, A. A., Koblan, L. W., Levy, J. M., et al. (2019). Search-and-replace genome editing without double-strand breaks or donor DNA. Nature, 576(7785), 149–157.
Matsoukas, I. G. (2020). Prime editing: Genome editing for rare genetic diseases without double-strand breaks or donor DNA. Frontiers in Genetics, 528.
Acknowledgments
M.E.W. would like to acknowledge the support of the Australian Research Council through Discovery Project Grants (DP200101860). M.E.W. holds a Fellowship from the Cancer Institute New South Wales (2021CDF1148). All figures were created with BioRender.com. We have been granted a license to use all figures in journal publications.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions
Author information
Authors and Affiliations
Contributions
ML and DMR equally contributed as first authors wrote the manuscript and repared the figures and tables. ML, DMR, SSM, AA, MM, SMJ, SF, MH, ML were involved in drafting the manuscript. MEW and MRA reviewed and provided suggessions for the manuscript. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
The authors have not disclosed any competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lotfi, M., Morshedi Rad, D., Mashhadi, S.S. et al. Recent Advances in CRISPR/Cas9 Delivery Approaches for Therapeutic Gene Editing of Stem Cells. Stem Cell Rev and Rep 19, 2576–2596 (2023). https://doi.org/10.1007/s12015-023-10585-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12015-023-10585-3