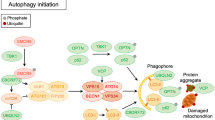
Abstract
Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disease that mainly affects the motor system. It is a very heterogeneous disorder, so far more than 40 genes have been described as responsible for ALS. The cause of motor neuron degeneration is not yet fully understood, but there is consensus in the literature that it is the result of a complex interplay of several pathogenic processes, which include alterations in nucleocytoplasmic transport, defects in transcription and splicing, altered formation and/or disassembly of stress granules and impaired proteostasis. These defects result in protein aggregation, impaired DNA repair, mitochondrial dysfunction and oxidative stress, neuroinflammation, impaired axonal transport, impaired vesicular transport, excitotoxicity, as well as impaired calcium influx. We argue here that all the above functions ultimately lead to defects in protein synthesis. Fused in Sarcoma (FUS) is one of the genes associated with ALS. It causes ALS type 6 when mutated and is found mislocalized to the cytoplasm in the motor neurons of sporadic ALS patients (without FUS mutations). In addition, FUS plays a role in all cellular functions that are impaired in degenerating motor neurons. Moreover, ALS patients with FUS mutations present the first symptoms significantly earlier than in other forms of the disease. Therefore, the aim of this review is to further discuss ALS6, detail the cellular functions of FUS, and suggest that the localization of FUS, as well as protein synthesis rates, could be hallmarks of the ALS phenotype and thus good therapeutic targets.
Graphical Abstract




Similar content being viewed by others
Data Availability
Data analysed during this study are included and refered in this published article.
References
Charcot, J. M. (1869). Deux cas d’atrophie musculaire progressive: avec leÌ sions de la substance grise et des faisceaux anteÌ rolateÌ raux de la moelle eÌ pinieÌ€re. Paris: Masson.
Cook, C., & Petrucelli, L. (2019). Genetic convergence brings clarity to the enigmatic Red line in ALS. Neuron, 101, 1057–1069.
Mathis, S., Goizet, C., Soulages, A., Vallat, J. M., & Masson, G. (2019). le. Genetics of amyotrophic lateral sclerosis: A review. J Neurol Sci [Internet]. Elsevier; [cited 2019 Apr 10];399:217–26. Available from: https://www.sciencedirect.com/science/article/pii/S0022510X19301017?via%3Dihub#bb0025
Petrov, D., Mansfield, C., Moussy, A., & Hermine, O. (2017). ALS clinical trials review: 20 years of failure. Are we any closer to registering a new treatment? Front Aging Neurosci (p. 68). Frontiers Research Foundation.
Hardiman, O., Al-Chalabi, A., Chio, A., Corr, E. M., Logroscino, G., Robberecht, W., et al. (2017). Amyotrophic lateral sclerosis. Nat Rev Dis Primers. Nature Publishing Group.
van Es, M. A., Hardiman, O., Chio, A., Al-Chalabi, A., Pasterkamp, R. J., Veldink, J. H., et al. (2017). Amyotrophic lateral sclerosis. The Lancet (pp. 2084–2098). Lancet Publishing Group.
van den Berg, L. H. (2014). Therapy of amyotrophic lateral sclerosis remains a challenge. Lancet Neurol (pp. 1062–1063). Lancet Publishing Group.
Van Damme, P., Robberecht, W., & Van Den Bosch, L. (2017). Modelling amyotrophic lateral sclerosis: Progress and possibilities. DMM Disease Models and Mechanisms (pp. 537–549). Company of Biologists Ltd.
Blair, I. P., Williams, K. L., Warraich, S. T., Durnall, J. C., Thoeng, A. D., Manavis, J., et al. (2010). FUS mutations in amyotrophic lateral sclerosis: clinical, pathological, neurophysiological and genetic analysis. J Neurol Neurosurg Psychiatry BMJ Publishing Group Ltd, 81, 639–645.
Kwiatkowski, T. J., Bosco, D. A., LeClerc, A. L., Tamrazian, E., Vanderburg, C. R., Russ, C., et al. (1979). Mutations in the FUS/TLS Gene on Chromosome 16 Cause Familial Amyotrophic Lateral Sclerosis. Science 2009;323:1205–8.
Vance, C., Rogelj, B., Hortobágyi, T., de Vos, K. J., Nishimura, A. L., Sreedharan, J., et al. (1979). Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 2009;323:1208–11.
Belzil, V. V., Valdmanis, P. N., Dion, P. A., Daoud, H., Kabashi, E., Noreau, A., et al. (2009). Mutations in FUS cause FALS and SALS in French and French Canadian populations. Neurology Lippincott Williams and Wilkins, 73, 1176–1179.
Bäumer, D., Hilton, D., Paine, S. M. L., Turner, M. R., Lowe, J., Talbot, K., et al. (2010). Juvenile ALS with basophilic inclusions is a FUS proteinopathy with FUS mutations. Neurology Lippincott Williams and Wilkins, 75, 611–618.
Huang, C., Zhou, H., Tong, J., Chen, H., Liu, Y. J., Wang, D., et al. (2011). FUS Transgenic Rats Develop the Phenotypes of Amyotrophic Lateral Sclerosis and Frontotemporal Lobar Degeneration. Cox GA, editor. PLoS Genet. Public Library of Science; ;7:e1002011.
Kiernan, M. C., Vucic, S., Cheah, B. C., Turner, M. R., Eisen, A., Hardiman, O., et al. (2011). Amyotrophic lateral sclerosis.The Lancet. p.942–55.
Gal, J., Zhang, J., Kwinter, D. M., Zhai, J., Jia, H., Jia, J., et al. (2011). Nuclear localization sequence of FUS and induction of stress granules by ALS mutants. Neurobiol Aging. ;32:2323.e27-2323.e40.
Reber, S., Stettler, J., Filosa, G., Colombo, M., Jutzi, D., Lenzken, S. C., et al. (2016). Minor intron splicing is regulated by FUS and affected by ALS-associated FUS mutants (35 vol., pp. 1504–1521). EMBO J. John Wiley & Sons, Ltd.
Sama, R. R., Ward, K., & Bosco, C. L. (2014). DA. Functions of FUS/TLS from DNA repair to stress response: implications for ALS. ASN Neuro: SAGE Publications.
Andersson, M. K., Ståhlberg, A., Arvidsson, Y., Olofsson, A., Semb, H., Stenman, G., et al. (2008). The multifunctional FUS, EWS and TAF15 proto-oncoproteins show cell type-specific expression patterns and involvement in cell spreading and stress response. Bmc Cell Biology, 9, 37.
Brelstaff, J., Lashley, T., Holton, J. L., Lees, A. J., Rossor, M. N., Bandopadhyay, R., et al. (2011). Transportin1: a marker of FTLD-FUS. Acta Neuropathologica, 122, 591–600.
Fujioka, Y., Ishigaki, S., Masuda, A., Iguchi, Y., Udagawa, T., Watanabe, H., et al. (2013). FUS-regulated region- and cell-type-specific transcriptome is associated with cell selectivity in ALS/FTLD. Sci Rep. Nature Publishing Group, 3, 2388.
Lagier-Tourenne, C., Polymenidou, M., Hutt, K. R., Vu, A. Q., Baughn, M., Huelga, S. C., et al. (2012). Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nature Neuroscience, 15, 1488–1497.
Mastrocola, A. S., Kim, S. H., Trinh, A. T., Rodenkirch, L. A., & Tibbetts, R. S. (2013). The RNA-binding protein fused in sarcoma (FUS) functions downstream of poly(ADP-ribose) polymerase (PARP) in response to DNA damage. Journal of Biological Chemistry, 288, 24731–24741.
Prpar Mihevc, S., Pavlin, M., Darovic, S., Živin, M., Podbregar, M., Rogelj, B., et al. (2017). Modelling FUS Mislocalisation in an In Vitro Model of Innervated Human Muscle. Journal of Molecular Neuroscience [Internet]. Springer New York LLC; [cited 2022 Dec 4];62:318–28. Available from: https://link.springer.com/article/10.1007/s12031-017-0940-y
Wächter, N., Storch, A., & Hermann, A. (2015). Human TDP-43 and FUS selectively affect motor neuron maturation and survival in a murine cell model of ALS by non-cell-autonomous mechanisms. Amyotroph Lateral Scler Frontotemporal Degener [Internet]. Amyotroph Lateral Scler Frontotemporal Degener; [cited 2022 Dec 4];16:431–41. Available from: https://pubmed.ncbi.nlm.nih.gov/26174443/
Jensen, B. K., McAvoy, K. J., Heinsinger, N. M., Lepore, A. C., Ilieva, H., Haeusler, A. R., et al. (2022). Targeting TNFα produced by astrocytes expressing amyotrophic lateral sclerosis-linked mutant fused in sarcoma prevents neurodegeneration and motor dysfunction in mice. Glia [Internet]. John Wiley & Sons, Ltd; [cited 2022 Dec 4];70:1426–49. Available from: https://onlinelibrary.wiley.com/doi/full/https://doi.org/10.1002/glia.24183
Guerrero, E. N., Wang, H., Mitra, J., Hegde, P. M., Stowell, S. E., Liachko, N. F., et al. (2016). TDP-43/FUS in motor neuron disease: Complexity and challenges.Prog Neurobiol.
Mitchell, J. C., McGoldrick, P., Vance, C., Hortobagyi, T., Sreedharan, J., Rogelj, B., et al. (2013). Overexpression of human wild-type FUS causes progressive motor neuron degeneration in an age- and dose-dependent fashion. Acta Neuropathol Springer, 125, 273–288.
Dichmann, D. S., & Harland, R. M. (2012). fus/TLS orchestrates splicing of developmental regulators during gastrulation. Genes dev (26 vol., pp. 1351–1363). Cold Spring Harbor Laboratory Press.
Hicks, G. G., Singh, N., Nashabi, A., Mai, S., Bozek, G., Klewes, L., et al. (2000). Fus deficiency in mice results in defective B-lymphocyte development and activation, high levels of chromosomal instability and perinatal death. Nature Genetics, 24, 175–179.
R, S. L., R, A., G, R. L., G, G. J., D.A.Q. M, F., G-HG-H, et al. (2014). PARP-1 dependent recruitment of the amyotrophic lateral sclerosis-associated protein FUS/TLS to sites of oxidative DNA damage. Nucleic Acids Research, 42, 307–314.
Wang, H., Guo, W., Mitra, J., Hegde, P. M., Vandoorne, T., Eckelmann, B. J., et al. (2018). Mutant FUS causes DNA ligation defects to inhibit oxidative damage repair in Amyotrophic Lateral Sclerosis. Nat Commun [Internet]. Nature Publishing Group; [cited 2019 Nov 3];9:3683. Available from: http://www.ncbi.nlm.nih.gov/pubmed/30206235
Kim, B. W., Jeong, Y. E., Wong, M., & Martin, L. J. (2020). DNA damage accumulates and responses are engaged in human ALS brain and spinal motor neurons and DNA repair is activatable in iPSC-derived motor neurons with SOD1 mutations.Acta Neuropathol Commun.
Mitra, J., Guerrero, E. N., Hegde, P. M., Liachko, N. F., Wang, H., Vasquez, V., et al. (2019). Motor neuron disease-associated loss of nuclear TDP-43 is linked to DNA double-strand break repair defects. Proc Natl Acad Sci U S A.
Walker, C., Herranz-Martin, S., Karyka, E., Liao, C., Lewis, K., Elsayed, W., et al. (2017). C9orf72 expansion disrupts ATM-mediated chromosomal break repair.Nat Neurosci.
Wang, W. Y., Pan, L., Su, S. C., Quinn, E. J., Sasaki, M., Jimenez, J. C., et al. (2013). Interaction of FUS and HDAC1 regulates DNA damage response and repair in neurons. Nature Neuroscience, 16, 1383–1391.
Higelin, J., Demestre, M., Putz, S., Delling, J. P., Jacob, C., Lutz, A. K., et al. (2016). FUS Mislocalization and Vulnerability to DNA Damage in ALS Patients Derived hiPSCs and Aging Motoneurons.Front Cell Neurosci. Frontiers Media SA; ;10.
Qiu, H., Lee, S., Shang, Y., Wang, W. Y., Au, K. F., Kamiya, S., et al. (2014). ALS-associated mutation FUS-R521C causes DNA damage and RNA splicing defects. Journal of Clinical Investigation, 124, 981–999.
Martinez-Macias, M. I., Moore, D. A., Green, R. L., Gomez-Herreros, F., Naumann, M., Hermann, A., et al. (2019). FUS (fused in sarcoma) is a component of the cellular response to topoisomerase I-induced DNA breakage and transcriptional stress.Life Sci Alliance. ;2.
Bennett, S. A., Tanaz, R., Cobos, S. N., Torrente, M. P., & Torrente Brooklyn, M. P. (2019 p). Epigenetics in amyotrophic lateral sclerosis: a role for histone post-translational modifications in neurodegenerative disease.Translational Research19–30.
Masala, A., Sanna, S., Esposito, S., Rassu, M., Galioto, M., Zinellu, A., et al. (2018). Epigenetic changes Associated with the expression of amyotrophic lateral sclerosis (ALS) causing genes. Neuroscience (390 vol., pp. 1–11). Elsevier Ltd.
Berson, A., Nativio, R., Berger, S. L., & Bonini, N. M. (2018). Epigenetic Regulation in Neurodegenerative Diseases.Trends Neurosci.
Cui, W., Yoneda, R., Ueda, N., & Kurokawa, R. (2018). Arginine methylation of translocated in liposarcoma (TLS) inhibits its binding to long noncoding RNA, abrogating TLS-mediated repression of CBP/p300 activity. Journal of Biological Chemistry. American Society for Biochemistry and Molecular Biology Inc.; ;293:10937–48.
Chen, K., Bennett, S. A., Rana, N., Yousuf, H., Said, M., Taaseen, S., et al. (2018). Neurodegenerative Disease Proteinopathies are connected to distinct histone post-translational modification landscapes. ACS Chem Neurosci American Chemical Society, 9, 938–948.
McClure, J. J., Li, X., & Chou, C. J. (2018). Advances and Challenges of HDAC Inhibitors in Cancer Therapeutics.Adv Cancer Res.
Ryu, H., Smith, K., Camelo, S. I., Carreras, I., Lee, J., Iglesias, A. H., et al. (2005). Sodium phenylbutyrate prolongs survival and regulates expression of anti-apoptotic genes in transgenic amyotrophic lateral sclerosis mice. J Neurochem. John Wiley & Sons, Ltd; ;93:1087–98.
Yoo, Y. E., & Ko, C. P. (2011). Treatment with trichostatin A initiated after disease onset delays disease progression and increases survival in a mouse model of amyotrophic lateral sclerosis. Exp Neurol Academic Press, 231, 147–159.
Guo, W., Naujock, M., Fumagalli, L., Vandoorne, T., Baatsen, P., Boon, R., et al. (2017). HDAC6 inhibition reverses axonal transport defects in motor neurons derived from FUS-ALS patients. Nat Commun Nature Publishing Group, 8, 861.
Rossaert, E., Pollari, E., Jaspers, T., Van Helleputte, L., Jarpe, M., Van Damme, P., et al. (2019). Restoration of histone acetylation ameliorates disease and metabolic abnormalities in a FUS mouse model. Acta Neuropathol Commun NLM (Medline), 7, 107.
Litt, M., Qiu, Y., & Huang, S. (2009). Histone arginine methylations: their roles in chromatin dynamics and transcriptional regulation (pp. 131–141). Biosci Rep. Portland Press.
Scaramuzzino, C., Monaghan, J., Milioto, C., Lanson, N. A., Maltare, A., Aggarwal, T., et al. (2013). Protein Arginine Methyltransferase 1 and 8 Interact with FUS to Modify Its Sub-Cellular Distribution and Toxicity In Vitro and In Vivo.PLoS One. ;8.
Tibshirani, M., Tradewell, M. L., Mattina, K. R., Minotti, S., Yang, W., Zhou, H., et al. Cytoplasmic sequestration of FUS/TLS associated with ALS alters histone marks through loss of nuclear protein arginine methyltransferase 1.
Ward, C. L., Boggio, K. J., Johnson, B. N., Boyd, J. B., Douthwright, S., Shaffer, S. A., et al. (2014). A loss of FUS/TLS function leads to impaired cellular proliferation.Cell Death Dis. Nature Publishing Group; ;5.
Rusk, N. (2008). When microRNAs activate translation. Nat Methods Nature Publishing Group, 5, 122–123.
Vasudevan, S., Tong, Y., & Steitz, J. A. (1979). Switching from Repression to Activation: MicroRNAs Can Up-Regulate Translation. Science 2007;318:1931–4.
Morlando, M., Dini Modigliani, S., Torrelli, G., Rosa, A., Di Carlo, V., Caffarelli, E., et al. (2012). FUS stimulates microRNA biogenesis by facilitating co-transcriptional Drosha recruitment. EMBO Journal, 31, 4502–4510.
De Santis, R., Santini, L., Colantoni, A., Peruzzi, G., de Turris, V., Alfano, V., et al. (2017). FUS Mutant Human Motoneurons Display altered transcriptome and microRNA pathways with implications for ALS Pathogenesis. Stem Cell Reports Cell Press, 9, 1450–1462.
Zhang, T., Wu, Y. C. C., Mullane, P., Ji, Y. J., Liu, H., He, L., et al. (2018). FUS regulates activity of MicroRNA-Mediated gene silencing. Mol Cell Cell Press, 69, 787–801e8.
Orozco, D., & Edbauer, D. (2013). FUS-mediated alternative splicing in the nervous system: Consequences for ALS and FTLD. J Mol Med. Springer; p. 1343–54.
Rogelj, B., Easton, L. E., Bogu, G. K., Stanton, L. W., Rot, G., Curk, T. T., et al. (2012). Widespread binding of FUS along nascent RNA regulates alternative splicing in the brain.Sci Rep. Nature Publishing Group; ;2.
Yu, Y., Chi, B., Xia, W., Gangopadhyay, J., Yamazaki, T., Winkelbauer-Hurt, M. E., et al. (2015). U1 snRNP is mislocalized in ALS patient fibroblasts bearing NLS mutations in FUS and is required for motor neuron outgrowth in zebrafish. Nucleic Acids Research, 43, 3208–3218.
Yu, Y., & Reed, R. (2015). FUS functions in coupling transcription to splicing by mediating an interaction between RNAP II and U1 snRNP. Proc Natl Acad Sci U S A National Academy of Sciences, 112, 8608–8613.
Ishigaki, S., Masuda, A., Fujioka, Y., Iguchi, Y., Katsuno, M., Shibata, A., et al. (2012). Position-dependent FUS-RNA interactions regulate alternative splicing events and transcriptions.Sci Rep. Nature Publishing Group; ;2.
Humphrey, J., Birsa, N., Milioto, C., Robaldo, D., Eberle, A. B., Kräuchi, R., et al. (2019). FUS ALS-causative mutations impact FUS autoregulation and the processing of RNA-binding proteins through intron retention. bioRxiv (p. 567735). Cold Spring Harbor Laboratory.
Grabowski, P. (2011). Alternative splicing takes shape during neuronal development.Curr Opin Genet Dev.
Tyzack, G. E., Luisier, R., Taha, D. M., Neeves, J., Modic, M., Mitchell, J. S., et al. (2019). Widespread FUS mislocalization is a molecular hallmark of amyotrophic lateral sclerosis.Brain.
Japtok, J., Lojewksi, X., Naumann, M., Klingenstein, M., Reinhardt, P., Sterneckert, J., et al. (2015). Stepwise acquirement of hallmark neuropathology in FUS-ALS iPSC models depends on mutation type and neuronal aging. Neurobiol Dis Academic Press Inc, 82, 420–429.
Marrone, L., Poser, I., Casci, I., Japtok, J., Reinhardt, P., Janosch, A., et al. (2018). Isogenic FUS-eGFP iPSC reporter lines enable quantification of FUS stress Granule Pathology that is rescued by drugs inducing Autophagy. Stem Cell Reports Elsevier, 10, 375–389.
Birsa, N., Bentham, M. P., & Fratta, P. (2020). Cytoplasmic functions of TDP-43 and FUS and their role in ALS. Semin Cell Dev Biol (pp. 193–201). Elsevier Ltd.
Stefl, R., Skrisovska, L., & Allain, F. H. T. (2005). RNA sequence- and shape-dependent recognition by proteins in the ribonucleoprotein particle.EMBO Rep.
Nostramo, R., Xing, S., Zhang, B., & Herman, P. K. (2019). Insights into the role of P-bodies and stress granules in protein quality control.Genetics.
Anderson, P., & Kedersha, N. (2008). Stress granules: the Tao of RNA triage.Trends Biochem Sci.
Lenzi, J., De Santis, R., De Turris, V., Morlando, M., Laneve, P., Calvo, A., et al. (2015). ALS mutant FUS proteins are recruited into stress granules in induced pluripotent stem cell-derived motoneurons. Company of Biologists Ltd, 8, 755–766.
Jain, S., Wheeler, J. R., Walters, R. W., Agrawal, A., Barsic, A., & Parker, R. (2016). ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure.Cell.
Bosco, D. A., Lemay, N., Ko, H. K., Zhou, H., Burke, C., Kwiatkowski, T. J., et al. (2010). Mutant FUS proteins that cause amyotrophic lateral sclerosis incorporate into stress granules.Hum Mol Genet.
Aulas, A., Stabile, S., Vande Velde, C., & Endogenous (2012). TDP-43, but not FUS, contributes to stress granule assembly via G3BP.Mol Neurodegener.
Baron, D. M., Kaushansky, L. J., Ward, C. L., Sama, R. R. K., Chian, R. J., Boggio, K. J., et al. (2013). Amyotrophic lateral sclerosis-linked FUS/TLS alters stress granule assembly and dynamics. Mol Neurodegener [Internet]. BioMed Central; [cited 2020 May 23];8:30. Available from: http://molecularneurodegeneration.biomedcentral.com/articles/https://doi.org/10.1186/1750-1326-8-30
Sama, R. R. K., Ward, C. L., Kaushansky, L. J., Lemay, N., Ishigaki, S., Urano, F., et al. (2013). FUS/TLS assembles into stress granules and is a prosurvival factor during hyperosmolar stress. Journal Of Cellular Physiology, 228, 2222–2231.
Dormann, D., Rodde, R., Edbauer, D., Bentmann, E., Fischer, I., Hruscha, A., et al. (2010). ALS-associated fused in sarcoma (FUS) mutations disrupt transportin-mediated nuclear import.EMBO Journal.
Sweeney, P., Park, H., Baumann, M., Dunlop, J., Frydman, J., Kopito, R., et al. (2017). Protein misfolding in neurodegenerative diseases: implications and strategies.Transl Neurodegener. Transl Neurodegener; ;6.
Zhang, P., Fan, B., Yang, P., Temirov, J., Messing, J., Kim, H. J., et al. (2019). Chronic optogenetic induction of stress granules is cytotoxic and reveals the evolution of ALS-FTD pathology (p. 8). Elife: eLife Sciences Publications Ltd.
Shelkovnikova, T. A., Robinson, H. K., Connor-Robson, N., & Buchman, V. L. (2013). Recruitment into stress granules prevents irreversible aggregation of FUS protein mislocalized to the cytoplasm. Cell Cycle Taylor and Francis Inc, 12, 3383–3391.
Alexander, E. J., Niaki, A. G., Zhang, T., Sarkar, J., Liu, Y., Nirujogi, R. S., et al. (2018). Ubiquilin 2 modulates ALS/FTD-linked FUS–RNA complex dynamics and stress granule formation (115 vol., pp. E11485–E11494). Proc Natl Acad Sci U S A. National Academy of Sciences.
Casci, I., Krishnamurthy, K., Kour, S., Tripathy, V., Ramesh, N., Anderson, E. N., et al. (2019). Muscleblind acts as a modifier of FUS toxicity by modulating stress granule dynamics and SMN localization.Nat Commun. Nature Research; ;10.
Ryu, H. H., Jun, M. H., Min, K. J., Jang, D. J., Lee, Y. S., Kim, H. K., et al. (2014). Autophagy regulates amyotrophic lateral sclerosis-linked fused in sarcoma-positive stress granules in neurons. Neurobiol Aging Elsevier Inc, 35, 2822–2831.
McCormick, C., & Khaperskyy, D. A. (2017). Translation inhibition and stress granules in the antiviral immune response.Nat Rev Immunol.
Limongi, D., & Baldelli, S. (2016). Redox Imbalance and Viral Infections in Neurodegenerative Diseases.Oxid Med Cell Longev.
Xue, Y. C., Feuer, R., Cashman, N., & Luo, H. (2018). Enteroviral infection: The forgotten link to amyotrophic lateral sclerosis?Front Mol Neurosci.
Celeste, D. B., & Miller, M. S. (2018). Reviewing the evidence for viruses as environmental risk factors for ALS: a new perspective. Cytokine (pp. 173–178). Academic Press.
Verma, A., & Berger, J. R. (2006). ALS syndrome in patients with HIV-1 infection.J Neurol Sci.
Shelkovnikova, T. A., An, H., Skelt, L., Tregoning, J. S., Humphreys, I. R., & Buchman, V. L. (2019). Antiviral Immune response as a trigger of FUS Proteinopathy in Amyotrophic lateral sclerosis. Cell Rep Elsevier B V, 29, 4496–4508e4.
Rhoads, S. N., Monahan, Z. T., Yee, D. S., & Shewmaker, F. P. (2018). The Role of Post-Translational Modifications on Prion-Like Aggregation and Liquid-Phase Separation of FUS. Int J Mol Sci. Multidisciplinary Digital Publishing Institute; p. 886.
Aulas, A., Velde, C., & Vande (2015). Alterations in stress granule dynamics driven by TDP-43 and FUS: A link to pathological inclusions in ALS? Front Cell Neurosci.Frontiers Research Foundation; ;9.
Zappulo, A., van den Bruck, D., Ciolli Mattioli, C., Franke, V., Imami, K., McShane, E., et al. (2017). RNA localization is a key determinant of neurite-enriched proteome. Nature Communications, 8, 583.
Eliscovich, C., & Singer, R. H. (2017). RNP transport in cell biology: the long and winding road.Curr Opin Cell Biol.
Jung, H., Yoon, B. C., & Holt, C. E. (2012). Axonal mRNA localization and local protein synthesis in nervous system assembly, maintenance and repair.Nat Rev Neurosci.
Karamyshev, A. L., & Karamysheva, Z. N. (2018). Lost in translation: Ribosome-associated mRNA and protein quality controls.Front Genet.
Kamelgarn, M., Chen, J., Kuang, L., Jin, H., Kasarskis, E. J., Zhu, H., & S A [Internet]. (2018). ALS mutations of FUS suppress protein translation and disrupt the regulation of nonsense-mediated decay. Proc Natl Acad Sci U. National Academy of Sciences; [cited 2019 Feb 15];115:E11904–13. Available from: http://www.ncbi.nlm.nih.gov/pubmed/30455313
López-Erauskin, J., Tadokoro, T., Baughn, M. W., Myers, B., McAlonis-Downes, M., Chillon-Marinas, C., et al. (2018). ALS/FTD-Linked Mutation in FUS Suppresses Intra-axonal Protein Synthesis and Drives Disease Without Nuclear Loss-of-Function of FUS. Neuron [Internet]. Cell Press; [cited 2019 Feb 15];100:816–830.e7. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0896627318308468
Nakaya, T., & Maragkakis, M. (2018). Amyotrophic lateral sclerosis associated FUS mutation shortens mitochondria and induces neurotoxicity. Sci Rep Nature Publishing Group, 8, 1–15.
de la Fuente, F. R., & Emc, F. (2020). FUS-ALS mutants alter FMRP phase separation equilibrium and impair protein translation Birsa N. bioRxiv [Internet]. Cold Spring Harbor Laboratory; [cited 2020 Sep 22];2020.09.14.296038. Available from: https://doi.org/10.1101/2020.09.14.296038
Bond, S., Lopez-Lloreda, C., Gannon, P. J., Akay-Espinoza, C., & Jordan-Sciutto, K. L. (2020). The integrated stress response and phosphorylated eukaryotic initiation factor 2α in neurodegeneration.J Neuropathol Exp Neurol.
Blum, J. A., & Gitler, A. D. (2022). Singling out motor neurons in the age of single-cell transcriptomics. Trends in Genetics Elsevier Current Trends, 38, 904–919.
Nichterwitz, S., Nijssen, J., Storvall, H., Schweingruber, C., Comley, L. H., Allodi, I., et al. (2020). LCM-seq reveals unique transcriptional adaptation mechanisms of resistant neurons and identifies protective pathways in spinal muscular atrophy. Genome Res [Internet]. Cold Spring Harbor Laboratory Press; [cited 2022 Dec 4];30:1083–96. Available from: https://pubmed.ncbi.nlm.nih.gov/32820007/
Lee, J., Yoo, M., & Choi, J. (2022). Recent advances in spatially resolved transcriptomics: challenges and opportunities. BMB Rep [Internet]. Korean Society for Biochemistry and Molecular Biology; [cited 2022 Dec 4];55:113. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8972138/
Russ, D. E., Cross, R. B. P., Li, L., Koch, S. C., Matson, K. J. E., Yadav, A., et al. (2021). A harmonized atlas of mouse spinal cord cell types and their spatial organization. Nature Communications 2021 12:1 [Internet]. Nature Publishing Group; [cited 2022 Dec 4];12:1–20. Available from: https://www.nature.com/articles/s41467-021-25125-1
Maniatis, S., Äijö, T., Vickovic, S., Braine, C., Kang, K., Mollbrink, A., et al. (2019). Spatiotemporal dynamics of molecular pathology in amyotrophic lateral sclerosis. Science [Internet]. Science; [cited 2022 Dec 4];364:89–93. Available from: https://pubmed.ncbi.nlm.nih.gov/30948552/
Kim, H. J. (2019). Cell fate control by translation: MRNA translation initiation as a therapeutic target for cancer development and stem cell fate control. Biomolecules. MDPI AG.
Powley, I. R., Kondrashov, A., Young, L. A., Dobbyn, H. C., Hill, K., Cannell, I. G., et al. (2009). Translational reprogramming following UVB irradiation is mediated by DNA-PKcs and allows selective recruitment to the polysomes of mRNAs encoding DNA repair enzymes. Genes dev (23 vol., pp. 1207–1220). Cold Spring Harbor Laboratory Press.
Bennetzen, M. V., Kosar, M., Bunkenborg, J., Payne, M. R., Bartkova, J., Lindström, M. S., et al. (2018). DNA damage-induced dynamic changes in abundance and cytosol-nuclear translocation of proteins involved in translational processes, metabolism, and autophagy (17 vol., pp. 2146–2163). Cell Cycle. Taylor and Francis Inc.
Nott, A., Le Hir, H., & Moore, M. J. (2004). Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes dev (18 vol., pp. 210–222). Cold Spring Harbor Laboratory Press.
Sephton, C. F., & Yu, G. (2015). The function of RNA-binding proteins at the synapse: implications for neurodegeneration. Cellular and Molecular Life Sciences.
Acknowledgements
We thank the people in the Foijer and Zatz labs for fruitful discussions.
Funding
This work was supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and an Abel Tasman fellowship to AA awarded by the University of Groningen. The funding agencies had no role in the design of the study and collection, analysis, and interpretation of data or in writing the manuscript.
Author information
Authors and Affiliations
Contributions
Amanda F. Assoni: Conceptualization; literature search; data curation; writing – original draft; writing – review and editing. Floris Foijer: Conceptualization; resources; data curation; writing – review and editing. Mayana Zatz: Conceptualization; supervision; resources; writing – review and editing.
Corresponding author
Ethics declarations
Ethics approval
Not Aplicable.
Consent for Publication
Not Aplicable.
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Assoni, A.F., Foijer, F. & Zatz, M. Amyotrophic Lateral Sclerosis, FUS and Protein Synthesis Defects. Stem Cell Rev and Rep 19, 625–638 (2023). https://doi.org/10.1007/s12015-022-10489-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12015-022-10489-8