Abstract
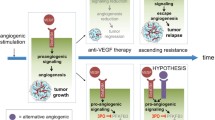
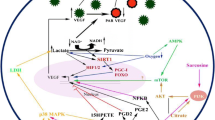
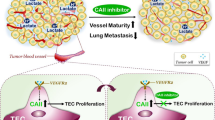
Metabolic status of the cells is important in the expression of the angiogenic phenotype in endothelial cells. Our earlier studies demonstrated the effects of metabolites such as lactate, citrate and lipoxygenase products, on VEGFA-VEGFR2 signaling pathway. Though this link between metabolite status and molecular mechanisms of angiogenesis is becoming evident, it is not clear how it affects genome-level expression in endothelial cells, critical to angiogenesis. In the present study, computational analysis was carried out on the transcriptome data of 4 different datasets where HUVECs were exposed to low and high glucose, both in vitro and in vivo, and the expression of a key enzyme involved in glucose metabolism is altered. The differentially expressed genes belonging to both VEGFA-VEGFR2 signaling pathway, as well as several VEGF signature genes as hub genes were also identified. These findings suggest the metabolite dependence, particularly glucose dependence, of angiogenesis, involving modulation of genome-level expression of angiogenesis- functional genome. This is important in tumor angiogenesis where reprogramming of metabolism is critical.









Similar content being viewed by others
Abbreviations
- Acronyms :
-
Expansion
- ADP:
-
Adenosine Di Phosphate
- AMPK:
-
AMP-activated protein kinase
- BAM:
-
Compressed Binary version of Sequence Alignment Map
- BC:
-
Betweenness Centrality
- BP:
-
Biological Processes
- CAM:
-
Cell Adhesion Molecule
- CC:
-
Closeness Centrality
- CNSA:
-
Chinese Nucleotide Sequence Archive
- DAVID:
-
Database for Annotation, Visualization and Integrated Discovery
- DC:
-
Degree Centrality
- DEG:
-
Differentially Expressed Gene
- EC:
-
Endothelial Cell
- ECM:
-
Extra Cellular Matrix
- ENA:
-
European Nucleotide Archive
- ER:
-
Endoplasmic Reticulum
- ERK:
-
Extracellular signal-Regulated Kinase
- ESCC:
-
Esophageal Squamous Cell Carcinoma
- FC:
-
Fold change
- FDR:
-
False Discovery Rate
- FOXO:
-
Forkhead Box
- FPKM:
-
Fragments Per Kilobase per Million Reads
- FTP:
-
File Transfer Protocol
- GEO:
-
Gene Expression Omnibus
- GO:
-
Gene Ontology
- HDF:
-
Human Dermal Fibroblast
- HDMEC:
-
Human Dermal Microvascular Endothelial Cell
- HIF:
-
Hypoxia Inducing Factor
- HISAT:
-
Hierarchical Indexing for Spliced Alignment of Transcripts
- HUVECs:
-
Human Umbilical Vein Endothelial Cells
- IRE1:
-
Inositol-Requiring Enzyme 1
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- MAPK:
-
Mitogen- Activated Protein Kinase
- MCODE:
-
Molecular Complex Detection
- NOD:
-
Nucleotide-binding and Oligomerization Domain
- PGC-1α:
-
Pparγ Coactivator 1 alpha
- PI3K-Akt:
-
Phosphatidyl-Inositol 3-Kinase- Ak strain Transforming
- PPARγ:
-
Peroxisome Proliferator- Activated Receptor gamma
- PPI:
-
Protein-Protein Interaction
- RIG-I:
-
Retinoic acid-Inducible Gene I
- RNA-seq:
-
RNA-sequencing
- SAM:
-
Sequence Alignment Map
- STRING:
-
Search Tool for the Retrieval of Interacting Genes/Proteins
- Wnt:
-
Wingless-related integration site
- Gene Symbol :
-
Gene Name
- ACAT2:
-
Acetyl-CoA Acetyltransferase 2
- ANLN:
-
Anillin Actin Binding Protein
- ASPM:
-
Assembly Factor for Spindle Microtubules
- ATF6:
-
Activating Transcription Factor 6
- C8orf33:
-
Chromosome 8 Open Reading Frame 33
- CALR:
-
Calreticulin
- CCL2:
-
C-C Motif Chemokine Ligand 2
- CCNB1:
-
Cyclin B1
- CCNB2:
-
Cyclin B2
- CEP55:
-
Centrosomal Protein 55
- CTSD:
-
Cathepsin D
- CXCL1:
-
C-X-C Motif Chemokine Ligand 1
- CXCL10:
-
C-X-C Motif Chemokine Ligand 10
- CXCL2:
-
C-X-C Motif Chemokine Ligand 2
- CXCL6:
-
C-X-C Motif Chemokine Ligand 6
- CXCL8:
-
C-X-C Motif Chemokine Ligand 8
- CXCR4:
-
C-X-C Motif Chemokine Receptor 4
- DNAJB9:
-
Dnaj Heat Shock Protein Family (Hsp40) Member B9
- FBXO5:
-
F-Box Protein 5
- FGF2:
-
Fibroblast Growth Factor 2
- HEK:
-
Human Epidermal Keratinocyte
- HERPUD1:
-
Homocysteine Inducible ER Protein with Ubiquitin Like Domain 1
- HIF:
-
Hypoxia Inducing Factor
- HYOU1:
-
Hypoxia Up-Regulated 1
- ICAM1:
-
Intercellular Adhesion Molecule 1
- IFI6:
-
Interferon Alpha Inducible Protein 6
- IFIH1:
-
Interferon Induced with Helicase C Domain 1
- IFIT3:
-
Interferon Induced Protein with Tetratricopeptide Repeats 3
- IFITM1:
-
Interferon Induced Transmembrane Protein 1
- ISG15:
-
ISG15 Ubiquitin Like Modifier
- LMNB1:
-
Lamin B1
- MDA5:
-
Melanoma Differentiation-Associated protein 5
- MRPS10:
-
Mitochondrial Ribosomal Protein S10
- MX1:
-
MX Dynamin Like GTPase 1
- MYCN:
-
MYCN Proto-Oncogene, bHLH Transcription Factor
- OAS1:
-
2′-5′-Oligoadenylate Synthetase 1
- OAS2:
-
2′-5′-Oligoadenylate Synthetase 2
- OAS3:
-
2′-5′-Oligoadenylate Synthetase 3
- P4HA2:
-
Prolyl 4-Hydroxylase Subunit Alpha 2
- P4HB:
-
Prolyl 4-Hydroxylase Subunit Beta
- PDIA6:
-
Protein Disulfide Isomerase Family A Member 6
- PFKFB3:
-
6-Phosphofructo-2-Kinase/Fructose-2,6-Biphosphatase 3
- PTGS1:
-
Prostaglandin-Endoperoxide Synthase 1
- RACGAP1:
-
Rac GTPase Activating Protein 1
- SDF2L1:
-
Stromal Cell Derived Factor 2 Like 1
- SELE:
-
Selectin E
- SLIT2:
-
Slit Guidance Ligand 2
- TGFB2:
-
Transforming Growth Factor Beta 2
- TNFAIP2:
-
TNF Alpha Induced Protein 2
- TPX2:
-
TPX2 Microtubule Nucleation Factor
- TYMP:
-
Thymidine Phosphorylase
- VCAM1:
-
Vascular Cell Adhesion Molecule 1
- VEGFA:
-
Vascular Endothelial Growth Factor A
- VEGFR2:
-
Vascular Endothelial Growth Factor Receptor 2
- XAF1:
-
XIAP Associated Factor 1
References
Carmeliet, P., & Jain, R. K. (2011). Molecular mechanisms and clinical applications of angiogenesis. Nature, 473(7347), 298–307. https://doi.org/10.1038/nature10144.
Chung, A. S., & Ferrara, N. (2011). Developmental and pathological angiogenesis. Annual Review of Cell and Developmental Biology, 27(1), 563–584. https://doi.org/10.1146/annurev-cellbio-092910-154002.
Sewduth, R., & Santoro, M. M. (2016). “Decoding” angiogenesis: New facets controlling endothelial cell behavior. Frontiers in Physiology, 7, 306 https://doi.org/10.3389/fphys.2016.00306.
Soumya, S. J., Athira, A. P., Binu, S., & Sudhakaran, P. R. (2016). mTOR as a Modulator of Metabolite Sensing Relevant to Angiogenesis, In Molecules to Medicine with mTOR: Translating Critical Pathways into Novel Therapeutic Strategies, 1st ed. (pp. 229–243). Kenneth Maiese: Academic Press. https://doi.org/10.1016/B978-0-12-802733-2.00014-1.
Kumar, V. B. S., Viji, R. I., Kiran, M. S., & Sudhakaran, P. R. (2007). Endothelial cell response to lactate: Implication of PAR modification of VEGF. Journal of Cellular Physiology, 211(2), 477–485. https://doi.org/10.1002/JCP.20955.
Binu, S., Soumya, S. J., & Sudhakaran, P. R. (2013). Metabolite control of angiogenesis: Angiogenic effect of citrate. The Journal of Physiology and Biochemistry, 69(3), 383–395. https://doi.org/10.1007/s13105-012-0220-9.
Soumya, S. J., Binu, S., Helen, A., Anil Kumar, K., Reddanna, P., & Sudhakaran, P. R. (2012). Effect of 15-lipoxygenase metabolites on angiogenesis: 15(S)-HPETE is angiostatic and 15(S)-HETE is angiogenic. The Journal of Inflammation Research, 61(7), 707–718. https://doi.org/10.1007/S00011-012-0463-5/.
Soumya, S. J., Binu, S., Helen, A., Reddanna, P., & Sudhakaran, P. R. (2013). 15 (S)-HETE-induced angiogenesis in adipose tissue is mediated through activation of PI3K/Akt/mTOR signaling pathway. Biochemistry and Cell Biology, 91(6), 498–505. https://doi.org/10.1139/BCB-2013-0037.
Soumya, S. J., Binu, S., Helen, A., Reddanna, P., & Sudhakaran, P. R. (2014). 15-LOX metabolites and angiogenesis: Angiostatic effect of 15(s)-hpete involves induction of apoptosis in adipose endothelial cells. PeerJ, 2, e635 https://doi.org/10.7717/peerj.635/.
Binu, S., Soumya, S. J., Kumar, V. B. S., & Sudhakaran, P. R. (2012). Poly-ADP-ribosylation of vascular endothelial growth factor and its implications on angiogenesis. Advances in Experimental Medicine and Biology, 749, 269–278. https://doi.org/10.1007/978-1-4614-3381-1_18.
Lau, A. N., & Vander Heiden, M. G. (2020). Metabolism in the tumor microenvironment. Annual Review of Cancer Biology, 4, 17–40. https://doi.org/10.1146/annurev-cancerbio-030419-033333.
Lin, X., Xiao, Z., Chen, T., Liang, S. H., & Guo, H. (2020). Glucose metabolism on tumor plasticity, diagnosis, and treatment. Frontiers Oncology, 10, 317 https://doi.org/10.3389/fonc.2020.00317.
Fadini, G. P., Albiero, M., Bonora, B. M., & Avogaro, A. (2019). Angiogenic abnormalities in diabetes mellitus: mechanistic and clinical aspects. The Journal of Clinical Endocrinology and Metabolism, 104(11), 5431–5444. https://doi.org/10.1210/JC.2019-00980.
Abhinand, C. S., Raju, R., Soumya, S. J., Arya, P. S., & Sudhakaran, P. R. (2016). VEGF-A/VEGFR2 signaling network in endothelial cells relevant to angiogenesis. The Journal of Cell Communication and Signaling, 10(4), 347–354. https://doi.org/10.1007/s12079-016-0352-8.
Sunitha, P., Raju, R., Sajil, C. K., Abhinand, C. S., Nair, A. S., Oommen, O. V., Sugunan, V. S., & Sudhakaran, P. R. (2019). Temporal VEGFA responsive genes in HUVECs: Gene signatures and potential ligands/receptors fine-tuning angiogenesis. Cell Communication and Signaling, 13(4), 561–571. https://doi.org/10.1007/S12079-019-00541-7/.
Bolger, A. M., Lohse, M., & Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Journal of Bioinformatics, 30(15), 2114–2120. https://doi.org/10.1093/bioinformatics/btu170.
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T., & Salzberg, S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nature Protocols, 11(9), 1650–1667. https://doi.org/10.1038/nprot.2016.095.
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., Marth, G., Abecasis, G., & Durbin, R. (2009). The sequence alignment/map format and SAMtools. Journal of Bioinformatics, 25(16), 2078–2079. https://doi.org/10.1093/bioinformatics/btp352.
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., & Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods, 5(7), 621–628. https://doi.org/10.1038/nmeth.1226.
Huang, D. W., Sherman, B. T., & Lempicki, R. A. (2009). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research, 37(1), 1–13. https://doi.org/10.1093/nar/gkn923.
Huang, D. W., Sherman, B. T., & Lempicki, R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols, 4(1), 44–57. https://doi.org/10.1038/nprot.2008.211.
Szklarczyk, D., Gable, A. L., Nastou, K. C., Lyon, D., Kirsch, R., Pyysalo, S., Doncheva, N. T., Legeay, M., Fang, T., Bork, P., Jensen, L. J., & von Mering, C. (2021). The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Research, 49(D1), D605–D612. https://doi.org/10.1093/nar/gkaa1074.
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., Amin, N., Schwikowski, B., & Ideker, T. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Research, 13(11), 2498–2504. https://doi.org/10.1101/GR.1239303.
Bader, G. D., & Hogue, C. W. V. (2003). An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics, 4(1), 1–27. https://doi.org/10.1186/1471-2105-4-2.
Tang, Y., Li, M., Wang, J., Pan, Y., & Wu, F. X. (2015). CytoNCA: A cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems, 127, 67–72. https://doi.org/10.1016/J.biosystems.2014.11.005.
Jeong, H., Mason, S. P., Barabási, A. L., & Oltvai, Z. N. (2001). Lethality and centrality in protein networks. Nature, 411(6833), 41–42. https://doi.org/10.1038/35075138.
Jin, G., Wang, Q., Pei, X., Li, X., Hu, X., Xu, E., & Li, M. (2019). mRNAs expression profiles of high glucose-induced memory in human umbilical vein endothelial cells. Diabetes, Metabolic Syndrome and Obesity: Targets and Therapy, 12, 1249–1261. https://doi.org/10.2147/DMSO.S206270.
Xu, E., Hu, X., Li, X., Jin, G., Zhuang, L., Wang, Q., & Pei, X. (2020). Analysis of long non-coding RNA expression profiles in high-glucose treated vascular endothelial cells. BMC Endocrine Disorders, 20(1), 1–10. https://doi.org/10.1186/S12902-020-00593-6/.
Zhang, S., Ke, Z., Yang, C., Zhou, P., Jiang, H., Chen, L., Li, Y., & Li, Q. (2021). High glucose causes distinct expression patterns of primary human skin cells by RNA sequencing. Frontiers in Endocrinology, 12, 152 https://doi.org/10.3389/fendo.2021.603645/.
Ambra, R., Manca, S., Palumbo, M. C., Leoni, G., Natarelli, L., de Marco, A., Consoli, A., Pandolfi, A., & Virgili, F. (2014). Transcriptome analysis of human primary endothelial cells (HUVEC) from umbilical cords of gestational diabetic mothers reveals candidate sites for an epigenetic modulation of specific gene expression. Genomics, 103(5–6), 337–348. https://doi.org/10.1016/j.ygeno.2014.03.003.
De Bock, K., Georgiadou, M., Schoors, S., Kuchnio, A., Wong, B. W., & Cantelmo, A. R. et al. (2013). Role of PFKFB3-driven glycolysis in vessel sprouting. Cell, 154(3), 651–663. https://doi.org/10.1016/j.cell.2013.06.037.
Du, W., Ren, L., Hamblin, M. H., & Fan, Y. (2021). Endothelial cell glucose metabolism and angiogenesis. Biomedicines, 9(2), 147 https://doi.org/10.3390/biomedicines9020147.
Kim, J. A., & Yeom, Y. I. (2018). Metabolic signaling to epigenetic alterations in cancer. Biomolecules & Therapeutics, 26(1), 69–80. https://doi.org/10.4062/biomolther.2017.185.
Zhou, J.-W., Wang, H., Sun, W., Han, N.-N., & Chen, L. (2020). ASPM is a predictor of overall survival and has therapeutic potential in endometrial cancer. The American Journal of Translational Research, 12(5), 1942–1953. https://pubmed.ncbi.nlm.nih.gov/32509189/.
Li, B., Zhu, H. B., Song, G. D., Cheng, J. H., Li, C. Z., Zhang, Y. Z., & Zhao, P. (2019). Regulating the CCNB1 gene can affect cell proliferation and apoptosis in pituitary adenomas and activate epithelial-to-mesenchymal transition. Oncology Letter, 18(5), 4651–4658. https://doi.org/10.3892/ol.2019.10847.
Fernández, L. P., Gómez de Cedrón, M., & Ramírez de Molina, A. (2020). Alterations of lipid metabolism in cancer: implications in prognosis and treatment. Frontier Oncology, 10, 2144 https://doi.org/10.3389/fonc.2020.577420.
Rojas, M. A., Santana, I., Lemtalsi, T., Caldwell, W., & Caldwell, R. B. (2020). Role of acyl-coenzyme A: cholesterol transferase (ACAT1) in pathological angiogenesis. Investigative Ophthalmology & Visual Science, 61(7), 5408
Pranjol, M. Z. I., Gutowski, N. J., Hannemann, M., & Whatmore, J. L. (2018). Cathepsin D non-proteolytically induces proliferation and migration in human omental microvascular endothelial cells via activation of the ERK1/2 and PI3K/AKT pathways. Biochimica et Biophysica Acta - Molecular and Cell Research, 1865(1), 25–33. https://doi.org/10.1016/j.bbamcr.2017.10.005.
Popson, S. A., & Hughes, C. C. W. (2010). A role for IFITM proteins in angiogenesis. FASEB Journal, 24(S1), 750–1. https://doi.org/10.1096/fasebj.24.1_supplement.750.1.
Popson, S. A., Ziegler, M. E., Chen, X., Holderfield, M. T., Shaaban, C. I., Fong, A. H., Welch-Reardon, K. M., Papkoff, J., & Hughes, C. C. W. (2014). Interferon-induced transmembrane protein 1 regulates endothelial lumen formation during angiogenesis. Arteriosclerosis, Thrombosis, and Vascular Biology, 34(5), 1011–1019. https://doi.org/10.1161/ATVBAHA.114.303352.
Cheriyath, V., Kaur, J., Davenport, A., Khalel, A., Chowdhury, N., & Gaddipati, L. (2018). G1P3 (IFI6), a mitochondrial localised antiapoptotic protein, promotes metastatic potential of breast cancer cells through mtROS. British Journal of Cancer, 119(1), 52–64. https://doi.org/10.1038/s41416-018-0137-3.
Liu, Z., Gu, S., Lu, T., Wu, K., Li, L., Dong, C., & Zhou, Y. (2020). IFI6 depletion inhibits esophageal squamous cell carcinoma progression through reactive oxygen species accumulation via mitochondrial dysfunction and endoplasmic reticulum stress. The Journal of Experimental & Clinical Cancer Research, 39(1), 1–28. https://doi.org/10.1186/s13046-020-01646-3.
Chawla-Sarkar, M., Lindner, D. J., Liu, Y. F., Williams, B. R., Sen, G. C., Silverman, R. H., & Borden, E. C. (2003). Apoptosis and interferons: role of interferon-stimulated genes as mediators of apoptosis. Apoptosis, 8(3), 237–249. https://doi.org/10.1023/A:1023668705040.
Maia, C. J., Rocha, S. M., Socorro, S., Schmitt, F., & Santos, C. R. (2016). Oligoadenylate synthetase 1 (OAS1) expression in human breast and prostate cancer cases, and its regulation by sex steroid hormones. Advances in Modern Oncology Research, 2(2), 97–110. https://doi.org/10.18282/amor.v2.i1.70.
Li, C., Wang, J., Zhang, H., Zhu, M., Chen, F., Hu, Y., Liu, H., Zhu, H., Li, C., Wang, J., Zhang, H., Zhu, M., Chen, F., Hu, Y., Liu, H., & Zhu, H. (2014). Interferon-stimulated gene 15 (ISG15) is a trigger for tumorigenesis and metastasis of hepatocellular carcinoma. Oncotarget, 5(18), 8429–8441. https://doi.org/10.18632/oncotarget.2316.
Zuo, C., Sheng, X., Ma, M., Xia, M., Ouyang, L., Zuo, C., Sheng, X., Ma, M., Xia, M., & Ouyang, L. (2016). ISG15 in the tumorigenesis and treatment of cancer: An emerging role in malignancies of the digestive system. Oncotarget, 7(45), 74393–74409. https://doi.org/10.18632/oncotarget.11911.
Aljohani, A. I., Joseph, C., Kurozumi, S., Mohammed, O. J., Miligy, I. M., Green, A. R., & Rakha, E. A. (2020). Myxovirus resistance 1 (MX1) is an independent predictor of poor outcome in invasive breast cancer. Breast Cancer Research and Treatment, 181(3), 541–551. https://doi.org/10.1007/S10549-020-05646-X.
Deng, C., Zhang, D., Shan, S., Wu, J., Yang, H., & Yu, Y. (2007). Angiogenic effect of intercellular adhesion molecule-1. Journal of Huazhong University of Science and Technology - Medical Science, 27(1), 9–12. https://doi.org/10.1007/S11596-007-0103-4.
Imai, S. I., & Guarente, L. (2014). NAD+ and sirtuins in aging and disease. Trends in Cell Biology, 24, 464–471. https://doi.org/10.1016/j.tcb.2014.04.002.
Hetz, C., & Papa, F. R. (2018). The unfolded protein response and cell fate control. Molecular Cell, 69, 169–181. https://doi.org/10.1016/j.molcel.2017.06.017.
Acknowledgements
The authors gratefully acknowledge the SIUCEB and DBT-BIF support to the Department of Computational Biology and Bioinformatics, University of Kerala, India for providing the necessary facilities and the Campus Computing Facility (CCF) at the Central Laboratory for Instrumentation and Facilitation (CLIF) at the University of Kerala for providing the HPC cluster facility to carry out this research work.
Author Contributions
Conceptualization, P.S. and P.R.S.; methodology, P.S. and P.R.S.; software, P.S.; validation, P.S. and K.R.A.; formal analysis, P.S. and P.R.S; data curation, P.S.; writing—original draft preparation, P.S. and P.R.S..; writing—review and editing, P.S., K.R.A., A.S.N., O.V.O. and P.R.S.; supervision, P.R.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Kerala State Council for Science, Technology and Environment (KSCSTE), Govt. of Kerala, by way of fellowship to K.R.A. P.R.S was supported by ISCA, Kolkata.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Sunitha, P., Arya, K.R., Nair, A.S. et al. Metabolite Effect on Angiogenesis: Insights from Transcriptome Analysis. Cell Biochem Biophys 80, 519–536 (2022). https://doi.org/10.1007/s12013-022-01078-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12013-022-01078-0