Abstract
The recently emerged SARS-like coronavirus (SARS-CoV-2) has continued to spread rapidly among humans with alarming upsurges in global mortality rates. A major key to tackling this virus is to disrupt its RNA replication process as previously reported for Remdesivir (Rem-P3). In this study, we theorize, using computational simulations, novel mechanisms that may underlie the binding of Rem-P3 to SARS-CoV-2 RdRp-NSPs complex; a multimeric assembly that drives viral RNA replication in human hosts. Findings revealed that while ATP-binding stabilized the replicative tripartite, Rem-P3 disintegrated the RdRp-NSP complex, starting with the detachment of the NSP7-NSP8 heterodimer followed by minimal displacement of the second NSP8 subunit (NSP8II). More so, Rem-P3 interacted with a relatively higher affinity (ΔGbind) while inducing high perturbations across the RdRp-NSP domains. D452, T556, V557, S682, and D760 were identified for their crucial roles in stacking the cyano-adenosine and 3,4-dihydroxyoxolan rings of Rem-P3 while its flexible P3 tail extended towards the palm domain blocking D618 and K798; a residue-pair identified for essential roles in RNA replication. However, ATP folded away from D618 indicative of a more coordinated binding favorable for nucleotide polymerization. We believe findings from this study will significantly contribute to the structure-based design of novel disruptors of the SARS-CoV-2 RNA replicative machinery.
Similar content being viewed by others
Introduction
Coronaviruses (CoVs) are a representative group of single-stranded, positive-sense RNA-viruses that are known to cause severe respiratory disorders, gastrointestinal and central nervous system (CNS) diseases in both humans and animals [1].
Severe acute respiratory syndrome CoV-2 is a newly emerged viral strain from the Coronaviridae family currently causing a global pandemic with severe threats to millions, and peradventure, billions of the world’s population. The middle east respiratory syndrome (MERS) CoV and severe acute respiratory syndrome CoV are other members of the family earlier reported, with mortality rates far lesser than SARS-CoV-2 [2,3,4]. Generically referred to as COVID-19, SARS-CoV-2 has recorded more mortality rates globally than MERS and SARS-CoV combined [5]. So far, deaths from SARS CoV-2 have been recently estimated at 207,000 with about 2.99 million infections [6]. Presently, there is no FDA-approved drug or vaccine for the treatment of SARS CoV-2, hence the highly urgent need to develop drugs and vaccines that will efficiently curtail its virulence.
SARS CoV-2 replication is very crucial to its pathogenesis even though quite complex [7]. Pivotal to the viral establishment in host cells is the assemblage of cytoplasmic and membrane-protected replication-transcription complexes (RTCs) [8, 9]. The RTCs most importantly coordinate the expression, replication, and amplification of the viral genome. They also create an environment for the virus to evade the hosts' immune system [10]. The production of viral sub-genomic mRNAs, synthesis of new genomic molecules, and template strand required for replication are hampered without the RTCs [11]. At the center of the RTCs is the RNA-dependent RNA polymerase (RdRp) subunit [10].
In the CoV genome, the 5′-terminus ORF1a and ORF1b frames encode polyprotein 1a and 1b which are cleaved into 16 non-structural proteins (NSPs) that cumulatively drive the replication and transcription phases of the virus [12,13,14,15,16].
Several viral cycles mediated by the different RdRp-NSPs lead to the full replication and translation of the viral genome. One of these steps is the nucleotide polymerization which involves at least three RdRp-NSPs namely NSP12, NSP7, and NSP8 [1].
The NSP12 is the core RdRp structure and it mimics the shape of a complete right hand with the palm, thumb, and finger domains [1, 17]. In addition, the NSP12 structure constitutes an N-terminal nidovirus RdRp-associated nucleotidyltransferase (NiRAN) domain common to all Nidovirales. This domain binds at the back-side of the C-terminal region of the RdRp and it is linked to the N-terminal domain by an interface domain [18]. Moreover, a beta-hairpin motif at the N-terminus binds at the interface located in between the palm and the NiRAN subdomains. Common to the coronavirus RdRp is the long-extended finger that forms a closed-ring structure upon an intersection with the thumb domain [18]. Among the seven classical catalytic motifs (A-E) of RdRp, five are situated within the palm domain which is the most conserved. The other two motifs F and G are located within the finger domain [19, 20].
The catalytic NSP12 subunit is highly essential for the viral replicative machinery and reportedly interacts with its cofactors; NSP7 and NSP8 which altogether drive genomic replication [1, 21]. Previous reports have emphasized that NSP12 is unable to perform its function as a single entity [22] but must exist in a tripartite complex with NSP7 and nsp8 for activating the replication of long RNAs which in turn increases template binding and processivity [21, 23]. A recent study revealed that the binding of NSP12 to template-primer RNA was dramatically increased by the presence of NSP7 and NSP8 [24]. NSP8 has the capability of initiating the replication process, which accounts for its description as a primase. This subunit can de novo synthesize about 6 nucleotides in length, which could serve as a primer for RNA synthesis by NSP12 RdRp. On the other hand, NSP7 is integral to the CoV replicase polyprotein which also functions as a primase and binds to NSP12 [21, 25, 26]. Together, the NSP7-NSP8 complex enhances the binding of NSP12 to RNA in addition to its enzymatic activity [7].
The functional multiunit complex of SARS-CoV-2 has been structurally resolved, containing one molecule of NSP12, two molecules of NSP8, and one molecule of NSP7. According to their findings, NSP12 polymerase was bound to a heterodimer of NSP7-NSP-8 while a second NSP8 subunit was bound at a distinct NSP12 site [24].
The binding of the NSP7-NSP8 heterodimer reportedly occurs above the NSP12 thumb subdomain and sandwiches the extended finger loops in between to attain conformational stability. This role is made possible by the bound heterodimeric NSP7 which makes most of the contact with the NSP12 while the first NSP8 molecule sparsely contacts the NSP12 subunit but rather interacts mainly with the NSP7. On the other hand, the second NSP8 subunit attaches to the top of the finger subdomain eliciting additional interactions with the interface domain [18].
Also, previous reports have shown that the complex demonstrated RNA polymerization activity on a poly-U template in the presence of adenosine triphosphate (ATP), which was inhibited effectively when active Remdesivir triphosphate was added [24].
Therefore, since this assembly is crucial for RNA synthesis, targeting it represents an important strategy to block the viral replication process. This could pave way for the design, synthesis, and repurposing of drugs that can disrupt this RdRp enzyme assembly, and consequently, viral replication.
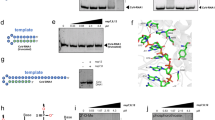
GS-5734/Remdesivir (Rem), a nucleoside analog, is a prodrug originally developed to combat Ebola virus, and functions by mimicking adenosine structure (Fig. 1) [27]. This drug reportedly converts into a hydrolyzed and tri-phosphorylated (active) metabolite (Rem-P3), a form that enhances its activity as a substrate for RdRp thereby replacing ATP. This event results in the termination of the polymerization process; regarded as ‘chain termination’ [27].
Efficacies of Rem in COVID-19 treatment have currently been evaluated in clinical trials, in the US and China [28], ever since the possibility was proposed in vitro. Rem exhibits broad-spectrum antiviral activities since most RNA viruses exhibit high structural similarities [27, 29].
Remdesivir has been experimentally reported to inhibit the replication of SARS-COV-2 [30], a feat that could facilitate its adoption and approval for COVID-19 treatment. Although, recent studies have reported its binding mode and interaction pattern in SARS-CoV-2, there is still room for additional structural insights with regards to its inhibitory dynamics and mechanisms, most especially, on how it disrupts the replicative machinery of SARS-CoV-2.
Therefore, in this study, we seek to unravel the structural modulatory mechanisms of Remdesivir on the SARS-CoV-2 NSP12-NSP8-NSP7 architecture, which is crucial for its replication in human host cells. Structural insights from this study would further corroborate existing details on the therapeutic functionality of Remdesivir in its active form. Using the recently resolved crystal structure of SARS-CoV-2 RdRp-NSP12-NSP8-NSP7, we modeled the differential binding of ATP and Rem-P3 and investigated structural events associated with the activities of both molecules using accelerated molecular dynamics (MD) simulations based on protocols used in our previous studies [31,32,33].
Conformational events were analyzed over the entire simulation period while relative binding modes of Rem-P3 and ATP were analyzed. We believe findings from this study would significantly contribute to the design of highly efficient inhibitors for disrupting SARS-CoV-2 replication in human hosts.
Computational methodology
Structural modeling of ligand and protein complexes
3D cryo-EM structures of unbound and Rem-P3-bound NSP12 assembly were obtained from the RCSB protein data bank with entries 7BV1 and 7BV2 [24]. These systems were prepared on the Graphic User Interface of UCSF Chimera to remove co-crystallized molecules not suitable for this study. In order to minimize computational cost, we employed the unbound NSP8-NSP12-NSP7-NSP8II RdRp assembly (without the template-RTP—7BV1) for our simulation runs, which was, however, superimposed with the template-RTP RdRp (7BV2) to determine the correct binding pocket and crystal orientation for Rem-P3. Magnesium ions (Mg2+) were retained in each NBP as they are reportedly essential for catalysis. This also applies to two zinc ions in the conserved metal-binding motifs made up of H295-C301-C306-C310 and C487-H642-C645-C646. 3D structure of ATP was retrieved from PubChem repository with entry CID5957. This was optimized structurally and geometrically at the B3LYP/6-311++G(d,p) theory level on the Gaussian16 program package [34] to obtain a stable conformation prior to molecular docking. A grid box was centered on Rem-P3 (in NSP12-NSP8-NSP7-NSP8II) and was used to obtain coordinates for the Rem-P3-bound NBP. ATP was then docked into the defined NBP using the Vina module [35]. Molecular docking yielded 10 ATP conformers among which the topmost ranked with a docking score of −5.9 kcal mol−1 was selected.
The best ATP binding pose with the highest score was selected and saved together with the NSP12-NSP8-NSP7-NSP8II to represent the ATP-bound NSP12 assembly.
GPU-accelerated molecular dynamics (MD) simulation
Altogether, the studied systems include: NSP8-NSP12-NSP7-NSP8-ATP, NSP8-NSP12-NSP7-NSP8-Rem-P3, and unbound NSP8-NSP12-NSP7-NSP8, which were subjected to total simulation runs of 600 ns. MD simulation was performed on Amber 18 Graphical Processor Unit [36] which enabled accelerated production runs. Parametrization of the individual receptor was performed on FF14SB forcefield. Also, Antechamber and Parmchk modules were used to generate .frcmod files for ATP and Rem-P3. The LEAP module was then used to generate topology and parameter files for the complexes in addition to system neutralization and explicit solvation [37]. These complexes were then minimized for 2500 steps with a 500 kcal/mol Å2 restraint potential and also for 5000 steps with no restraints. Simultaneous heating (0–300k) and equilibration steps were performed followed by production runs that were restarted subsequently. The resulting trajectories were saved at every 1 ns after which they were analyzed with the integrated CPPTRAJ module [38].
Relative stabilities of the SARS-CoV-2 multimers were determined by measuring the Cα-root mean square deviations (RMSDs) while other metrics such as the root mean square fluctuation (RMSF), the radius of gyration (RoG), and solvent accessibility surface area (SASA) were used to measure per-residual motions, structural compactness, and solvent-surface motions [31,32,33, 39,40,41].
Trajectorial motions of the molecules were also projected along with two principal components (PC1 and PC2) for insights into their dynamics and motion patterns [42,43,44]. Data plots and snapshots for time-based visual analysis were obtained using the Origin analytical software and UCSF Chimera GUI [45], Biovia Discovery Studio [46], and GIMP 2.0.
Binding free energy calculation
The molecular mechanics/Poisson Boltzmann surface area (MM/PBSA) method [47] was used to measure the binding of ATP and Rem-P3 at the NSP12-nucleoside pocket of the SARS-CoV-2 multimer (NSP8-NSP12-NSP7-NSP8II). Estimated ΔGbinds were decomposed into various energy components [ΔEele = electrostatic energy; ΔEvdW = van der Waals energy; ΔGsolv = solvation free energy ΔGgas = gas-phase free energy; ΔGGB = polar desolvation energy; −TΔS = entropy, ΔEnp,sol = non-polar solvation energy] based on the expression: ΔGbind = ΔEvdW + ΔEele + ΔEGB + ΔEnp,sol – TΔS to further understand the differential binding mechanisms of the compounds. Also, we tracked the interaction energies of the compounds over the simulation period to gain insights into the binding dynamics. Per-residue energies were also decomposed to identify residues that significantly contribute to inhibitor/substrate binding and stability at the NSP12 active pocket.
Results and discussion
Remdesivir systematically disintegrates the SARS-CoV-2 RdRp assembly
To understand the possible effects of Rem-P3 and ATP on the SARS-CoV-2 NSP complex, snapshots were taken from the resulting trajectories at various time-frames as the simulation proceeded. Comparative visual analysis was then carried out for the unbound, Rem-P3- and ATP-bound systems.
We observed that the RdRp-NSP multimer remained as a unit in the unbound and ATP-bound systems from the initial to the final frames while dissimilar structural occurrences were observed in the Rem-P3-bound complex (Fig. 2).
Time-based conformational sampling revealed that the RdRp-NSP assembly was intact in the presence of Rem-P3 until around 150 ns when the NSP8 subunit was firstly detached from its NSP7 counterpart into the solvent environment. This was then followed by the displacement of the NSP7 subunit from the loop interface of the NSP12-thumb domain at the next 20 ns time-frame (170 ns). Besides, we observed that the second NSP8 subunit was minimally displaced from its binding interface on NSP12. The disintegrated RdRp-NSP complex was maintained until the end of the simulation (Fig. 3).
Based on previous studies involving SARS-CoV, NSP7, and NSP8 play important roles in de novo initiation, primer extension, RNA synthesis, and replication, particularly when complexed with NSP12 [21, 48, 49]. Also, an assemblage of the NSP7-NSP8 heterodimer with NSP12 reportedly enhanced its RNA binding and polymerase activities [7, 21].
We further investigated the effects of both compounds on the integrity of the NSP12 multimer over the simulation period by comparing parameters for the unbound, ATP-bound, and Rem-P3-bound systems. Firstly, we estimated the stability of the complexes using the RMSD metrics and the result plotted in Fig. 4a. As shown, the unbound and ATP-bound complexes were stable from the beginning to the end of the simulations. On the contrary, the Rem-P3 -bound NSP complex was structurally stable (RMSD < 2 Å) until ~140 ns where high instability was observed, characterized by an unusual spike in Cα motions after which the system was re-stabilized until the end of the run.
Conformational analyses for whole structural and nucleoside site stability using Cα-RMSD and RoG parameters. a Shows large structural instability for Rem-P3 system. b Finally equilibrated (FE)-RMSD using more stable time-frames (170–200 ns). c Active site stability—FE-RMSD. d FE-RoG plot showing the degree of active site compactness
This observation could correlate with the disintegration of the NSP7-NSP8 heterodimer from the NSP12 polymerase as reported above (Fig. 3) thereby explaining high conformational perturbations induced by Rem-P3 when bound to the SARS-CoV-2 RdRp-NSP complex.
A possible explanation is that the disintegration of the NSP7-NSP8 heterodimer could further destabilize the NSP12 subunit, which could, in turn, affect binding interfaces for other NSPs necessary for RNA replication [1, 21, 49]. Another interesting and important finding was that the ATP-bound NSP-complex was more stable than the unbound system.
This could imply that the binding of ATP to NSP12 further enhanced the integrity of the SARS-CoV-2 NSP8-NSP12-NSP7-NSP8 complex. To minimize entropic effects, we defined finally equilibrated RMSDs (FE-RMSDs) from the ultimate time frames (170–200 ns) where the systems stabilized. As seen in Fig. 4b, huge discrepancies exist between the stabilities of the simulated complexes, with a high FE-RMSD peak for the Rem-P3 bound system. This could correlate with the systematic disintegration of the RdRp-NSP complex induced by Rem-P3 relative to ATP which rather stabilized the complex.
Estimated mean FE-RMSDs of the systems were in the order ATP < unbound < Rem-P3 as presented in Supplementary Table 1. These stable time-frames were then utilized for subsequent conformational analyses.
Also, we projected the motions of the RdRp-NSP complexes along with two principal components (PC1 vs. PC2). From the PCA plot in Supplementary Fig. S1, we observed similar directions of motions among the unbound and ATP-bound RdRp-NSPs while a more dispersed motion pattern was observed for the Rem-P3-bound complex.
We further monitored systemic fluctuations among constituent residues of each NSP subunits using the FE-RMSF metrics, derived from the stable time-frames. Our findings revealed that per-residual motions were relatively high in the Rem-P3-bound NSP complex compared to ATP-bound and unbound systems (Supplementary Fig. S2). Collectively, the mean FE-RMSF was lowest in the ATP-system further indicative of the importance of ATP in stabilizing the RdRp-NSP multimeric complex. Estimated mean FE-RMSFs of the systems also followed in the ATP < Unbound < Rem-P3 order.
We then mapped out the distinct subunits of the RdRp-NSP complex (including NSP12 sub-domains) and measured their fluctuations with respect to the binding of the compounds.
Our findings further emphasized the perturbative effects of Rem-P3 across the NSP8-NSP12-NSP7-NSP8 assembly. The disruptive effects of Rem-P3 binding were most pronounced in the NSP7-bound NSP8 heterodimer (FE-RMSF = 41.6 Å) and could have led to their detachment from the finger loop interfaces, which was also highly perturbed (FE-RMSF = 9.0 Å) according to our calculations (Fig. 5).
Cumulatively, high Cα motions induced by Rem-P3 at the NSP12 subdomains affected the binding and stability of associated NSPs at the interfaces (Supplementary Table 2). More so, high fluctuations induced by Rem-P3 among residues of the NSP12 N-terminal and finger subdomains could have disrupted interface interactions with the second NSP8 subunit (NSP8II), which also exhibited high structural perturbations (FE-RMSF = 11.9 Å, 9.0 Å).
Interestingly, the NSP12 subdomains, in addition to other NSPs (7 and 8) exhibited minimal residual motions in the presence of ATP, indicative of the stability of the NSP complex, even compared to the unbound system. Hence, we can presume, that the binding of ATP further enhanced structural integrity across the RdRp-NSP assembly.
Differential stabilities of the NSP12-nucleoside sites were further evaluated, and as observed (Fig. 4c), the Rem-P3-bound nucleoside site was more unstable compared to the ATP-bound and unbound sites. This could suggest a possible correlation between the activities of Rem-P3 at the nucleoside pocket and its disruptive effects on the entire RdRp-NSP complex.
Also, Rem-P3 induced high Ca motions at the nucleoside site as further estimated using the FE-RoG metrics, which is also an indicator of structural compactness [41]. Relatively, the nucleoside side was more compact when bound by ATP, even compared to the unbound NSP12-pocket (Fig. 4d).
Furthermore, we projected trajectorial motions of the compounds at the active pockets with respect to their binding activities. From the PCA plot, we could observe that Rem-P3 exhibited highly unstable motions at the nucleoside pocket while ATP was coordinately bound with more compact motions indicative of its systemic stability.
Complementarily, we masked the compounds for FE-RMSD calculations to further determine their binding stability. As shown in Fig. 6, Rem-P3 was highly unstable (FE-RMSD = 3.5 Å ± 0.5) while ATP demonstrated stable motions at the binding pocket (FE-RMSD = 1.5 Å ± 0.8).
Analysis of Rem-P3 and ATP motions, and orientations at the active site region of RdRp-NSP12. a Ligand FE-RMSD. b Binding modes of Rem-P3 and (d). ATP at the hydrophobic NSP12 pocket. Interacting residues are also shown in addition to the degree of hydrophobicity color-coded from least → highly hydrophobic; −3.00 → 3.00. c PCA clustering of ligand motions over the simulation period
Also, we observed that ATP assumed a folded conformation at the hydrophobic pocket of the NSP12 subunit, which could have accounted for the compact site architecture (Fig. 6d). On the contrary, Rem-P3 exhibited an extended conformation with its cyano-adenosine ring stacked in between a visible hydrophobic patch (Fig. 6b). The effect of this Rem-P3 binding mode could be seen in the pocket which was less compact.
Using average structures for the Rem-P3- and ATP-bound complexes, we closely analyzed relative orientations at the NSP12-nucleoside site and complementary interactions (Fig. 7). As mentioned above, ATP ultimately assumed a compact conformation, with an inwardly folded P3 tail while Rem-P3 rather exhibited a linearly stretched conformation. As observed, its cyano-nucleotide portion extended into the pocket created by D452, T556, V557, T680, S681, and S682 (finger domain).
This ring orientation could allow for high-affinity (NH–O) interactions with D452 and T556, which could, in turn, hold the cyano-adenosine ring in place. More so, its P3 tail extends into the palm domain towards K798, D618, K621, R553, and D623. This orientation enables the incorporation of its monophosphate group into the primer strand as resolved by a recent structural study [24]. Also, the 3,4 dihydroxyoxolan ring was coordinated by N691, S759, and D760, which constitute the catalytic active center. These findings, altogether, indicate an orientation that potentiates its inhibitory activities on the RNA elongation process. Relatively, we could observe that the P3 tail of ATP was folded away from D618 and K798, an important residue-pair responsible SARS-CoV RdRp polymerase activity as previously reported [50]. The relevance of these respective residues to catalytic RdRp activity and Rem-P3 blockade has been recently reported [24].
Binding free energy calculations and decomposition
The extent to which the compounds bind to the RdRp-NSP complex was determined using the MM/PBSA method [51, 52]. Stable time-frames (170–200 ns) were also selected to minimize entropical effects and our results are presented in Table 1. Accordingly, Rem-P3 was strongly bound than ATP with ∆Gbind difference of −13.2 kcal mol−1.
This relatively higher affinity of Rem-P3 for the NSP12 nucleoside site further reflects its ability to competitively impede and replace the natural substrate [53]. This high-affinity, as estimated, could also potentiate its covalent-binding activities as recently reported [24].
Analysis of the binding components further revealed that the binding of Rem-P3 was more unfavorable in the polar region as evidenced by higher a ∆Gnp,sol value. This could suggest that Rem-P3 does not only block ATP binding but binds deeply in the hydrophobic nucleoside pocket of the NSP12 subunit.
This could favor its retention and involvement with more residues of the pocket. Also, vdW contributions to Rem-P3 binding were relatively higher, which may compensate for electrostatic (∆Eele) effects that were negated by unfavorable polar solvation energies (∆Gsolv). Cumulatively, ∆EvdW and ∆Eele highly favored gas-phase interactions (∆Ggas) of Rem-P3 relative to ATP.
Also, we calculated the energies of individual residues and their respective contributions to the disparate binding of both compounds. Energy contributions >−1 kcal mol−1 were considered favorable and results are presented in Fig. 8.
As shown, total energy contributions to Rem-P3 were highly favorable for D452 (−4.1 kcal mol−1), D760 (−3.6 kcal mol−1), S682 (−2.8 kcal mol−1), R555 (−2.0 kcal mol−1), T556 (−1.6 kcal mol−1), T687 (−1.1 kcal mol−1), V557 (−1.0 kcal mol−1) and T680 (−1.1 kcal mol−1) in that order.
This could further emphasize the importance of D452 in enhancing Rem-P3 binding via interaction with its cyano-adenosine portion, and D760 via interactions with its 3,4 dihydroxyoxolan ring (Fig. 7a). Relative to ATP, most of these energies were reduced and highly unfavorable. For instance, D425 and R553 contributed unfavorable energies (+ve) of 1.0 and 5.3 kcal mol−1 to ATP which could minimize its interaction affinity compared to Rem-P3. However, S549 (−1.1 kcal mol−1), R555 (−5.9 kcal mol−1), K621 (−2.5 kcal mol−1), and D623 (−2.5 kcal mol−1) all contributed favorably to ATP binding.
The roles of D618 and K798 to the binding of Rem-P3 were further defined with electrostatic contributions of −11.4 and −1.2 kcal mol−1, respectively. On the contrary, electrostatic effects by D618 were highly unfavorable for ATP (10.1 kcal mol−1).
This could suggest that ATP does not interfere with the roles mediated by D618 during nucleotide polymerization [50]. More so, while D452 electrostatically favored Rem-P3 binding (−5.3 kcal mol−1), an unfavorable contribution of +12.5 kcal mol−1 was estimated for ATP. Similar Rem-P3 → ATP (∆Eele) transitions were observed for T556 (−5.4 → 1.9 kcal mol−1), D623 (−10.1 → 9.0 kcal mol−1), D760 (−11.2 → 9.2 kcal mol−1), and D761 (−8.3 → 7.4 kcal mol−1). Presumably, residues that favored Rem-P3 binding at the nucleotide site of RdRp-NSP12 rather contributed unfavorably to ATP binding, which could limit ATP affinity in the region. Also, in Rem-P3, unfavorable ∆Eele contributions were highly compensated for by per-residue ∆EvdW energies which were favorable in R555 (−2.9 kcal mol−1), T556 (−1.4 kcal mol−1), V557 (1.0 kcal mol−1), D623 (−2.3 kcal mol−1), R624 (−1.4 kcal mol−1), T680 (−1.1 kcal mol−1), S681 (−1.4 kcal mol−1) and S682 (−2.8 kcal mol−1). These were decreased to −1.7 kcal mol−1 (R555), −0.4 kcal mol−1 (T556), +0.4 kcal mol−1 (D623), and −0.7 kcal mol−1 (R624) in the ATP-complex.
Taken together, we could presume that Rem-P3 binds uncoordinatedly at the NSP12 nucleoside site, interacting with a high number of residues along its path. This binding pattern favorably enhanced its affinity and, perhaps, longer retention at the NSP12 site until it exerts maximum destabilizing effects sufficient to disrupt the RdRp-NSP multimeric assembly.
Relatively, the ability of ATP to fold its flexible P3 tail away from D618 and K798 in a compact conformation indicates a selective and coordinated binding, favorable for the nucleotide polymerization process.
Conclusion
In this study, we investigated the differential binding of Rem-P3 and ATP to the RdRp-NSP assembly. This involved subunit binding of an NSP7-NSP8 heterodimer to NSP12 while a second NSP8 (NSP8II) was bound at a distant site. Rem-P3-, ATP- and unbound complexes were subjected to GPU-accelerated molecular dynamics simulation of 600 ns after which the trajectories were sampled conformationally. Findings revealed that the RdRp-NSP complex was more stable in the presence of ATP when compared to the unbound system while on the other hand, Rem-P3 appeared to disintegrate the complex. The NSP7-NSP8 heterodimer was firstly detached at ~150 ns from the NSP12 while the second NSP8 (NSP8II) subunit was seen to be minimally displaced at the ultimate time-frame. Conformational analysis performed with RMSD and RoG parameters further revealed that Rem-P3 induced a high degree of structural instability compared to the ATP and unbound system. RMSF analysis revealed relatively higher subdomain perturbations in the Rem-P3-bound RdRp-NSP complex. Also, analysis of ligand motions revealed that ATP exhibited a more stable and compact motion compared to Rem-P3, which could have also reflected on the active site, which was less deviated and more compact in the ATP-system. Analysis of binding dynamics revealed that Rem-P3 exhibited an extended conformation that allowed it to interact with D542, T556, V557, T680, S681, and S682 of the NSP12-finger domain via its cyano-adenosine ring while its 3,4 dihydroxyoxolan ring oriented towards D760 and N961 forming high-affinity H-bonds. Also, its P3 tail was extended towards the palm domain, interacting with D618 and K798. This binding pattern clearly interferes with the roles mediated by D618 and K798 in the RNA replication process. On the contrary, ATP exhibited a more selective and coordinated binding wherein it assumed a folded (compact) conformation away from these residues. A binding energy value of −33.4 kcal mol−1 was estimated for Rem-P3 while ATP had a ΔGbind estimate of −20.5 kcal mol−1. This further reflected the ability of Rem-P3 to competitively replace ATP at the NSP12-nucleoside pocket. Per-residue energy decomposition further emphasized the roles of D542, T556, V557, T680, S681, S682, D618, and K798 to the high-affinity binding of Rem-P3. We believe findings from this study will significantly contribute to drug design endeavors, particularly structure-based approaches. This could enable the further design of novel compounds that can bind selectively and strongly to the NSP12-nucleoside site and disrupt interactions with other NSPs.
References
Kirchdoerfer, R. N., & Ward, A. B. (2019). Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nature Communications. https://doi.org/10.1038/s41467-019-10280-3.
Yang, D., & Leibowitz, J. L. (2015). The structure and functions of coronavirus genomic 3′ and 5′ ends. Virus Research. https://doi.org/10.1016/j.virusres.2015.02.025.
Drosten, C., Günther, S., Preiser, W., Van der Werf, S., Brodt, H. R., Becker, S., & Doerr, H. W. (2003). Identification of a novel coronavirus in patients with severe acute respiratory syndrome. New England Journal of Medicine, 348(20), 1967–1976. https://doi.org/10.1056/NEJMoa030747.
Zaki, A. M., Van Boheemen, S., Bestebroer, T. M., Osterhaus, A. D. M. E., & Fouchier, R. A. M. (2012). Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. New England Journal of Medicine, 367(19), 1814–1820. https://doi.org/10.1056/NEJMoa1211721.
Mahase, E. (2020). Coronavirus: covid-19 has killed more people than SARS and MERS combined, despite lower case fatality rate, 641(February), 2020. https://doi.org/10.1136/bmj.m641.
Dong, E., Du, H., & Gardner, L. (2020). An interactive web-based dashboard to track COVID-19 in real time. The Lancet Infectious Diseases. https://doi.org/10.1016/S1473-3099(20)30120-1.
Imbert, I., Guillemot, J. C., Bourhis, J. M., Bussetta, C., Coutard, B., Egloff, M. P., & Canard, B. (2006). A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO Journal, 25(20), 4933–4942. https://doi.org/10.1038/sj.emboj.7601368.
Ahlquist, P., Noueiry, A. O., Lee, W.-M., Kushner, D. B., & Dye, B. T. (2003). Host factors in positive-strand RNA virus genome replication. Journal of Virology, 77(15), 8181–8186. https://doi.org/10.1128/jvi.77.15.8181-8186.2003.
Miller, W. A., & Koev, G. (2000). Synthesis of subgenomic RNAs by positive-strand RNA viruses. Virology. https://doi.org/10.1006/viro.2000.0421.
van Hemert, M. J., van den Worm, S. H. E., Knoops, K., Mommaas, A. M., Gorbalenya, A. E., & Snijder, E. J. (2008). SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathogens, 4(5), e1000054. https://doi.org/10.1371/journal.ppat.1000054.
Ziebuhr, J. (2005). The coronavirus replicase. Current topics in microbiology and immunology. Berlin, Heidelberg: Springer. https://doi.org/10.1007/3-540-26765-4_3.
Mackenzie, J. (2005). Wrapping things up about virus RNA replication. Traffic. https://doi.org/10.1111/j.1600-0854.2005.00339.x.
Miller, S., & Krijnse-Locker, J. (2008). Modification of intracellular membrane structures for virus replication. Nature Reviews Microbiology. https://doi.org/10.1038/nrmicro1890.
Salonen, A., Ahola, T., & Kääriäinen, L. (2004). Viral RNA replication in association with cellular membranes. Current Topics in Microbiology and Immunology. https://doi.org/10.1007/3-540-26764-6_5.
Zumla, A., Chan, J. F. W., Azhar, E. I., Hui, D. S. C., & Yuen, K. Y. (2016). Coronaviruses-drug discovery and therapeutic options. Nature Reviews Drug Discovery. https://doi.org/10.1038/nrd.2015.37.
Snijder, E. J., Decroly, E., & Ziebuhr, J. (2016). The nonstructural proteins directing coronavirus RNA synthesis and processing. Advances in Virus Research. https://doi.org/10.1016/bs.aivir.2016.08.008.
Gao, Y., Yan, L., Huang, Y., Liu, F., Zhao, Y., Cao, L., Rao, Z. (2020). Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. https://doi.org/10.1126/science.abb7498.
Peng, Q., Peng, R., Yuan, B., Zhao, J., Wang, M., Wang, X., Shi, Y. (2020). Structural and biochemical characterization of the nsp12-nsp7-nsp8 core polymerase complex from SARS-CoV-2. Cell Reports. https://doi.org/10.1016/j.celrep.2020.107774.
Gorbalenya, A. E., Pringle, F. M., Zeddam, J. L., Luke, B. T., Cameron, C. E., Kalmakoff, J., Ward, V. K. (2002). The palm subdomain-based active site is internally permuted in viral RNA-dependent RNA polymerases of an ancient lineage. Journal of Molecular Biology. https://doi.org/10.1016/S0022-2836(02)01033-1.
Venkataraman, S., Prasad, B. V. L. S., & Selvarajan, R. (2018). RNA dependent RNA polymerases: Insights from structure, function and evolution. Viruses. https://doi.org/10.3390/v10020076.
Subissi, L., Posthuma, C. C., Collet, A., Zevenhoven-Dobbe, J. C., Gorbalenya, A. E., Decroly, E., & Imbert, I. (2014). One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proceedings of the National Academy of Sciences of the United States of America, 111(37), E3900–E3909. https://doi.org/10.1073/pnas.1323705111.
Ahn, D. G., Choi, J. K., Taylor, D. R., & Oh, J. W. (2012). Biochemical characterization of a recombinant SARS coronavirus nsp12 RNA-dependent RNA polymerase capable of copying viral RNA templates. Archives of Virology, 157(11), 2095–2104. https://doi.org/10.1007/s00705-012-1404-x.
Te Velthuis, A. J. W., Van Den Worm, S. H. E., & Snijder, E. J. (2012). The SARS-coronavirus nsp7 + nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Research. https://doi.org/10.1093/nar/gkr893.
Yin, W., Mao, C., Luan, X., Shen, D. D., Shen, Q., Su, H., Xu, H. E. (2020). Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by Remdesivir. Science. https://doi.org/10.1126/science.abc1560.
Sevajol, M., Subissi, L., Decroly, E., Canard, B., & Imbert, I. (2014). Insights into RNA synthesis, capping, and proofreading mechanisms of SARS-coronavirus. Virus Research, 194, 90–99. https://doi.org/10.1016/j.virusres.2014.10.008.
Lehmann, K. C., Gulyaeva, A., Zevenhoven-Dobbe, J. C., Janssen, G. M. C., Ruben, M., Overkleeft, H. S., & Gorbalenya, A. E. (2015). Discovery of an essential nucleotidylating activity associated with a newly delineated conserved domain in the RNA polymerase-containing protein of all nidoviruses. Nucleic Acids Research, 43(17), 8416–8434. https://doi.org/10.1093/nar/gkv838.
Tchesnokov, E. P., Feng, J. Y., Porter, D. P., & Götte, M. (2019). Mechanism of inhibition of ebola virus RNA-dependent RNA polymerase by remdesivir. Viruses, 11(4), 1–16. https://doi.org/10.3390/v11040326.
Rodriguez-Torres, M., Yoshida, E. M., Marcellin, P., Srinivasan, S., Purohit, V. S., Wang, C., & Hammond, J. L. (2014). A phase 2 study of filibuvir in combination with pegylated IFN alfa and ribavirin for chronic HCV. Annals of Hepatology. https://doi.org/10.1016/s1665-2681(19)30843-9.
Weber, C., & Opatz, T. (2019). Bisbenzylisoquinoline alkaloids. Alkaloids: Chemistry and Biology. https://doi.org/10.1016/bs.alkal.2018.07.001.
Wang, M., Cao, R., Zhang, L., Yang, X., Liu, J., Xu, M., Xiao, G. (2020). Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Research, 2019–2021. https://doi.org/10.1038/s41422-020-0282-0.
Olotu, F. A., Munsamy, G., & Soliman, M. E. S. (2018). Does size really matter? Probing the efficacy of structural reduction in the optimization of bioderived compounds—a computational “proof-of-concept”. Computational and Structural Biotechnology Journal, 16, 573–586. https://doi.org/10.1016/J.CSBJ.2018.11.005.
Abdullahi, M., Olotu, F. A., & Soliman, M. E. (2018). Solving the riddle: unraveling the mechanisms of blocking the binding of leukotoxin by therapeutic antagonists in periodontal diseases. Journal of Cellular Biochemistry, 119(11). https://doi.org/10.1002/jcb.27254.
Olotu, F. A., Agoni, C., Adeniji, E., Abdullahi, M., & Soliman, M. E. (2018). Probing gallate-mediated selectivity and high-affinity binding of epigallocatechin gallate: a way-forward in the design of selective inhibitors for anti-apoptotic Bcl-2 proteins. Applied Biochemistry and Biotechnology. https://doi.org/10.1007/s12010-018-2863-7.
Frisch, M. J. G., Trucks, W., Schlegel, H. B., Scuseria, G. E., Robb, M. A., Cheeseman, J. R., Sonnenberg, J. L. (2016). Gaussian 16., Inc.: Wallingford, CT. https://doi.org/10.1007/s12231-009-9086-3.
Trott, O., & Olson, A. (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. Journal of Computational Chemistry, 31(2), 455–461. https://doi.org/10.1002/jcc.21334.AutoDock.
Case, D. A. (2018). Amber 18. San Francisco: University of California.
Case, D. A., Cheatham, T. E., Darden, T., Gohlke, H. & & Luo, R. et al. (2005). The Amber biomolecular simulation programs. Journal of Computational Chemistry, 26, 1668–1688.
Roe, D. R., & Cheatham, T. E. (2013). PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. Journal of Chemical Theory and Computation, 9(7), 3084–3095. https://doi.org/10.1021/ct400341p.
Olotu, F. A., & Soliman, M. E. S. (2018). From mutational inactivation to aberrant gain-of-function: Unraveling the structural basis of mutant p53 oncogenic transition. Journal of Cellular Biochemistry, 119(3). https://doi.org/10.1002/jcb.26430.
Ali, S., Hassan, M., Islam, A., & Ahmad, F. (2014). A review of methods available to estimate solvent-accessible surface areas of soluble proteins in the folded and unfolded states. Current Protein & Peptide Science, 15(5), 456–476. https://doi.org/10.2174/1389203715666140327114232.
Lobanov, M. Y., Bogatyreva, N. S., & Galzitskaya, O. V. (2008). Radius of gyration as an indicator of protein structure compactness. Molecular Biology, 42(4), 623–628. https://doi.org/10.1134/S0026893308040195.
Sittel, F., Jain, A., & Stock, G. (2014). Principal component analysis of molecular dynamics: On the use of Cartesian vs. internal coordinates. Journal of Chemical Physics, 141(1). https://doi.org/10.1063/1.4885338.
David, C. C., & Jacobs, D. J. (2014). Principal component analysis: a method for determining the essential dynamics of proteins. Methods in Molecular Biology, 1084, 193–226. https://doi.org/10.1007/978-1-62703-658-0_11.
Lawal, M., Olotu, F. A., & Soliman, M. E. S. (2018). Across the blood-brain barrier: neurotherapeutic screening and characterization of naringenin as a novel CRMP-2 inhibitor in the treatment of Alzheimer’s disease using bioinformatics and computational tools. Computers in Biology and Medicine, 98, 168–177. https://doi.org/10.1016/J.COMPBIOMED.2018.05.012.
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., & Ferrin, T. E. (2004). UCSF Chimera? A visualization system for exploratory research and analysis. Journal of Computational Chemistry, 25(13), 1605–1612. https://doi.org/10.1002/jcc.20084.
BIOVIA, D. S. (2015). Discovery studio modeling environment. San Diego: Dassault Systèmes.
Ylilauri, M., & Pentikäinen, O. T. (2013). MMGBSA as a tool to understand the binding affinities of filamin-peptide interactions. Journal of Chemical Information and Modeling, 53(10), 2626–2633. https://doi.org/10.1021/ci4002475.
Brockway, S. M., Clay, C. T., Lu, X. T., & Denison, M. R. (2003). Characterization of the expression, intracellular localization, and replication complex association of the putative mouse hepatitis virus RNA-dependent RNA polymerase. Journal of Virology. https://doi.org/10.1128/jvi.77.19.10515-10527.2003.
von Brunn, A., Teepe, C., Simpson, J. C., Pepperkok, R., Friedel, C. C., Zimmer, R., Haas, J. (2007). Analysis of intraviral protein–protein interactions of the SARS coronavirus ORFeome. PLoS ONE. https://doi.org/10.1371/journal.pone.0000459.
te Velthuis, A. J. W., Arnold, J. J., Cameron, C. E., van den Worm, S. H. E., & Snijder, E. J. (2009). The RNA polymerase activity of SARS-coronavirus nsp12 is primer dependent. Nucleic Acids Research. https://doi.org/10.1093/nar/gkp904.
Wang, C., Greene, D., Xiao, L., Qi, R., & Luo, R. (2018). Recent developments and applications of the MMPBSA method. Frontiers in Molecular Biosciences, 4. https://doi.org/10.3389/fmolb.2017.00087.
Akher, F. B., Farrokhzadeh, A., Olotu, F. A., Agoni, C., & Soliman, M. E. S. (2019). Biomolecular Chemistry, 1176–1190. https://doi.org/10.1039/c8ob02811g.
Vliegen, I., Paeshuyse, J., Zhong, W., & Neyts, J. (2015). In vitro combinations containing Tegobuvir are highly efficient in curing cells from HCV replicon and in delaying/preventing the development of drug resistance. Antiviral Research. https://doi.org/10.1016/j.antiviral.2015.05.011.
Acknowledgements
The authors would like to thank the Center for High-Performance Computing (CHPC), Cape Town, South Africa, for making computational resources available.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Olotu, F.A., Omolabi, K.F. & Soliman, M.E.S. Piece of the puzzle: Remdesivir disassembles the multimeric SARS-CoV-2 RNA-dependent RNA polymerase complex. Cell Biochem Biophys 79, 175–187 (2021). https://doi.org/10.1007/s12013-021-00977-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12013-021-00977-y