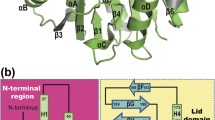
Abstract
Among the known covalent damages that can occur spontaneously to proteins, the formation of isoaspartyl linkages through deamidation of asparagines and isomerization of aspartates may be one of the most rapid forms under conditions of physiological pH and temperature. The protein l-isoaspartyl methyltransferase (PIMT) is thought to recognize l-isoaspartyl residues and repair this kind of damaged proteins. Curiously, there is a potential functional difference between bacterial and mammalian PIMTs. Herein, we present the crystal structure of Escherichia coli PIMT (EcPIMT) at a resolution of 1.8 Å. The enzyme we investigated was able to remain bound to its product S-adenosylhomocysteine (SAH) during crystallization. Analysis indicates that the high affinity of EcPIMT for SAH might lead to the lower activity of the enzyme.



Similar content being viewed by others
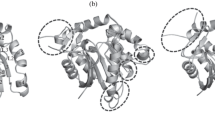
References
Reissner, K. J., & Aswad, D. W. (2003). Deamidation and isoaspartate formation in proteins: Unwanted alterations or surreptitious signals? Cellular and Molecular Life Sciences, 60(7), 1281–1295.
Perna, A. F., Ingrosso, D., De Santo, N. G., Galletti, P., & Zappia, V. (1995). Mechanism of erythrocyte accumulation of methylation inhibitor S-adenosylhomocysteine in uremia. Kidney International, 47(1), 247–253.
Shirasawa, T., Endoh, R., Zeng, Y. X., Sakamoto, K., & Mori, H. (1995). Protein l-isoaspartyl methyltransferase: Developmentally regulated gene expression and protein localization in the central nervous system of aged rat. Neuroscience Letters, 188(1), 37–40.
Yamamoto, A., Takagi, H., Kitamura, D., Tatsuoka, H., Nakano, H., Kawano, H., et al. (1998). Deficiency in protein l-isoaspartyl methyltransferase results in a fatal progressive epilepsy. Journal of Neuroscience, 18(6), 2063–2074.
Kondo, T., Shirasawa, T., Itoyama, Y., & Mori, H. (1996). Embryonic genes expressed in Alzheimer’s disease brains. Neuroscience Letters, 209(3), 157–160.
David, C. L., Keener, J., & Aswad, D. W. (1999). Isoaspartate in ribosomal protein S11 of Escherichia coli. Journal of Bacteriology, 181(9), 2872–2877.
Kindrachuk, J., Parent, J., Davies, G. F., Dinsmore, M., Attah-Poku, S., & Napper, S. (2003). Overexpression of l-isoaspartate O-methyltransferase in Escherichia coli increases heat shock survival by a mechanism independent of methyltransferase activity. Journal of Biological Chemistry, 278(51), 50880–50886.
Kern, R., Malki, A., Abdallah, J., Liebart, J. C., Dubucs, C., Yu, M. H., et al. (2005). Protein isoaspartate methyltransferase is a multicopy suppressor of protein aggregation in Escherichia coli. Journal of Bacteriology, 187(4), 1377–1383.
Powell, H. R. (1999). The Rossmann Fourier autoindexing algorithm in MOSFLM. Acta Crystallographica D: Biological Crystallography, 55(Pt 10), 1690–1695.
Vagin, A., & Teplyakov, A. (1997). Molrep: An automated program for molecular replacement. Journal of Applied Crystallography, 30, 1022–1025.
Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., et al. (1998). Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallographica D: Biological Crystallography, 54(Pt 5), 905–921.
Murshudov, G., Vagin, A., & Dodson, E. (1997). Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallographica D: Biological Crystallography, 53(Pt 3), 240–255.
Winn, M. D., Isupov, M. N., & Murshudov, G. N. (2001). Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallographica D: Biological Crystallography, 57(Pt 1), 122–133.
Emsley, P., & Cowtan, K. (2004). Coot: Model-building tools for molecular graphics. Acta Crystallographica D: Biological Crystallography, 60(Pt 12), 2126–2132.
Laskowski, R. A., MacArthur, M. W., Moss, D. S., & Thornton, J. M. (1993). PROCHECK: A program to check the stereochemical quality of protein structures. Journal of Applied Crystallography, 26, 283–291.
Smith, C. D., Carson, M., Friedman, A. M., Skinner, M. M., Delucas, L., Chantalat, L., et al. (2002). Crystal structure of human l-isoaspartyl-O-methyl-transferase with S-adenosyl homocysteine at 1.6-Å resolution and modeling of an isoaspartyl-containing peptide at the active site. Protein Science, 11(3), 625–635.
Tanaka, Y., Tsumoto, K., Yasutake, Y., Umetsu, M., Yao, M., Fukada, H., et al. (2004). How oligomerization contributes to the thermostability of an archaeon protein. Protein l-isoaspartyl-O-methyltransferase from Sulfolobus tokodaii. Journal of Biological Chemistry, 279(31), 32957–32967.
Bennett, E. J., Bjerregaard, J., Knapp, J. E., Chavous, D. A., Friedman, A. M., Royer, W. E., Jr., et al. (2003). Catalytic implications from the Drosophila protein l-isoaspartyl methyltransferase structure and site-directed mutagenesis. Biochemistry, 42(44), 12844–12853.
Wallace, A. C., Laskowski, R. A., & Thornton, J. M. (1995). LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Engineering, 8(2), 127–134.
Griffith, S. C., Sawaya, M. R., Boutz, D. R., Thapar, N., Katz, J. E., Clarke, S., et al. (2001). Crystal structure of a protein repair methyltransferase from Pyrococcus furiosus with its l-isoaspartyl peptide substrate. Journal of Molecular Biology, 313(5), 1103–1116.
Acknowledgments
Financial support for this project was provided by research grants from the Chinese National Natural Science Foundation (grant nos. 30025012 and 10979039), the Chinese Ministry of Science and Technology (grant nos. 2006CB806500, 2006CB910200, and 2006AA02A318), the Chinese Academy of Sciences (grant no. KSCX2-YW-R-60) and the Chinese Ministry of Education (grant no. 20070358025), Anhui Provincial Natural Science Foundation (grant no. 090413081). We thank Yi Zhu and Xu Cao for their assistance in the plasmid construction.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Fang, P., Li, X., Wang, J. et al. Crystal Structure of the Protein l-Isoaspartyl Methyltransferase from Escherichia coli . Cell Biochem Biophys 58, 163–167 (2010). https://doi.org/10.1007/s12013-010-9103-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12013-010-9103-2