Abstract
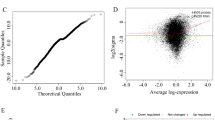
Exposure to hexavalent chromium [Cr(VI)] causes human and animal hepatotoxicity. However, it is unclear how Cr(VI) induces hepatotoxicity, nor is it clear which pathways and genes may be involved. This study aimed to identify the key molecular pathways and genes engaged in Cr(VI)-induced hepatotoxicity. Publicly available microarray GSE19662 was downloaded from the Gene Expression Omnibus database. GSE19662 consists of primary rat hepatocyte (PRH) groups treated with or without 0.10 ppm potassium dichromate (PD), with three samples per group. Compared to the control group, a total of 400 differentially expressed genes were obtained. Specially 262 and 138 genes were up- and downregulated in PD-treated PRHs, respectively. Gene ontology (GO) enrichment indicated that those DEGs were primarily engaged in many biological processes, including androgen biosynthetic process, the positive regulation of cell death, the response to activity, the toxic substance and hepatocyte growth factor stimulus, and others. Kyoto Encyclopedia of Genes and Genomes (KEGG) suggested that the DEGs are fundamentally enriched in hepatocellular carcinoma (HCC), hepatitis B, p53, PI3K-Akt, MAPK, AMPK, metabolic pathways, estrogen, cGMP-PKG, metabolic pathways, etc. Moreover, many genes, including UBE2C, TOP2A, PRC1, CENPF, and MKI67, might contribute to Cr(VI)-induced hepatotoxicity. Taken together, this study enhances our understanding of the regulation, prevention, and treatment strategies of Cr(VI)-induced hepatotoxicity.







Similar content being viewed by others
References
Mishra S, Bharagava RN (2016) Toxic and genotoxic effects of hexavalent chromium in environment and its bioremediation strategies. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev 34:1–32. https://doi.org/10.1080/10590501.2015.1096883
Patlolla C, Barnes C, Yedjou V, Velma P (2009) Oxidative stress, DNA damage, and antioxidant enzyme activity induced by hexavalent chromium in Sprague-Dawley rats. Environ Toxicol 24:66–73. https://doi.org/10.1002/tox.20395
Navya K, Kumar GP, Chandrasekhar Y, Anilakumar KR (2018) Evaluation of potassium dichromate (K2Cr2O7)-induced liver oxidative stress and ameliorative effect of Picrorhiza kurroa extract in Wistar albino rats. Biol Trace Elem Res 184(1):154–164. https://doi.org/10.1007/s12011-017-1172-2
Saha R, Nandi R, Saha B (2011) Sources and toxicity of hexavalent chromium. J Coord Chem 64(10):1782–1806. https://doi.org/10.1080/00958972.2011.583646
Ni XM, Wan L, Liang PP, Zheng RP, Lin ZY, Chen RC, Pei MK, Shen YJ (2020) The acute toxic effects of hexavalent chromium on the liver of marine medaka (Oryzias melastigma). Comp Biochem Physiol C Toxicol Pharmacol 231:108734. https://doi.org/10.1016/j.cbpc.2020.108734
Yang QY, Han B, Li SY, Wang XQ, Wu PF, Liu Y, Li JY, Han BQ, Deng N, Zhang ZG (2022) The link between deacetylation and hepatotoxicity induced by exposure to hexavalent chromium. J Adv R 35:129–140. https://doi.org/10.1016/j.jare.2021.04.002
El-Demerdash FM, El-Sayed RA, Abdel-Daim MM (2021) Hepatoprotective potential of Rosmarinus officinalis essential oil against hexavalent chromium-induced hematotoxicity, biochemical, histological, and immunohistochemical changes in male rats. Environ Sci Pollut Res 28(14):17445–17456. https://doi.org/10.1007/s11356-020-12126-8
Zhao Y, Yan J, A-Pei L, Z-Li Z, Z-Run L, K-Jun G, K-Chi Z, Ruan Q, Guo L (2019) Bone marrow mesenchymal stem cells could reduce the toxic effects of hexavalent chromium on the liver by decreasing endoplasmic reticulum stress-mediated apoptosis via SIRT1/HIF-1 signaling pathway in rats. Toxicol Lett 310:31–38. https://doi.org/10.1016/j.toxlet.2019.04.007
Liu XT, Rehman MU, Mehmood K, Huang SC, Tian XX, Wu XX, Zhou DH (2018) Ameliorative effects of nano-elemental selenium against hexavalent chromium-induced apoptosis in broiler liver. Environ Sci Pollut Res Int 25(16):15609–15615. https://doi.org/10.1007/s11356-018-1758-z
Yang QY, Han B, Xue JD, Lv YY, Li SY, Liu Y, Wu PF, Wang XQ, Zhang ZG (2020) Hexavalent chromium induces mitochondrial dynamics disorder in rat liver by inhibiting AMPK/PGC-1a signaling pathway. Environ Pollut 265:114855. https://doi.org/10.1016/j.envpol.2020.114855
Luo M, Huang SC, Zhang JL, Zhang LH, Mehmood K, Jiang JH, Zhang NY, Zhou DH (2019) Effect of selenium nanoparticles against abnormal fatty acid metabolism induced by hexavalent chromium in chicken’s liver. Environ Sci Pollut Res Int 26(21):21828–21834. https://doi.org/10.1007/s11356-019-05397-3
Yan JY, Huang HR, Liu ZP, Shen JY, Ni J, Han JW, Wang RJ, Lin DR, Hu BW, Jin LF (2020) Hedgehog signaling pathway regulates hexavalent chromium-induced liver fibrosis by activation of hepatic stellate cells. Toxicol Lett 320:1–8. https://doi.org/10.1016/j.toxlet.2019.11.017
Deng YP, Johnson DR, Guan X, Ang CY, Ai JM, Perkins EJ (2010) In vitro gene regulatory networks predict in vivo function of liver. BMC Syst Biol 4:153. https://doi.org/10.1186/1752-0509-4-153
Xiong Y, Lu J, Fang QL, Lu YY, Xie CR, Wu HT, Yin ZY (2019) UBE2C functions as a potential oncogene by enhancing cell proliferation, migration, invasion, and drug resistance in HCC cells. Bioscience Rep 39(4): BSR20182384. 10.1042/ BSR20182384
Zhu MQ, Wu MN, Bian SY, Song QQ, Xiao MB, Huang H, You L, Zhang JP, Zhang J, Cheng C, Ni WK, Zheng WJ (2021) DNA primase subunit 1 deteriorated progression of HCC by activating AKT/mTOR signaling and UBE2C-mediated p53 ubiquitination. Cell Biosci 11(1):42. https://doi.org/10.1186/s13578-021-00555-y
Yang ZQ, Wu XL, Li JB, Zheng Q, Niu JW, Li SW (2021) CCNB2, CDC20, AURKA, TOP2A, MELK, NCAPG, KIF20A, UBE2C, PRC1, and ASPM may be potential therapeutic targets for HCC using integrated bioinformatic analysis. Int J Gen Med 14:10185–10194. https://doi.org/10.2147/IJGM.S341379
Si TF, Huang ZL, Jiang YH, Walker-Jacobs A, Gill S, Hegarty R, Hamza M, Khorsandi SE, Jassem W, Heaton N, Ma Y (2021) Expression levels of three key genes CCNB1, CDC20, and CENPF in HCC are associated with antitumor immunity. Front Oncol 11:738841. https://doi.org/10.3389/fonc.2021.738841
Wu SY, Liao P, Yan LY, Zhao QY, Xie ZY, Dong J, Sun HT (2021) Correlation of MKI67 with prognosis, immune infiltration, and T cell exhaustion in HCC. BMC gastroenterol 21(1):416. https://doi.org/10.1186/s12876-021-01984-2
Li L, Wei JR, Song Y, Fang S, Du YY, Li Z, Zeng TT, Zhu YH, Li Y, Guan XY (2021) TROAP switches DYRK1 activity to drive HCC progression. Cell Death Dis 12(1):125. https://doi.org/10.1038/s41419-021-03422-3
Su LS, Zhang GH, Kong XD (2021) A Novel five-gene signature for prognosis prediction in HCC. Front Oncol 11:642563. https://doi.org/10.3389/fonc.2021.642563
Zheng YC, Shi Y, Yu S, Han YY, Kang K, Xu HF, Gu HJ, Sang XT, Chen Y, Wang JY (2019) GTSE1, CDC20, PCNA, and MCM6 synergistically affect regulations in cell cycle and indicate poor prognosis in liver cancer. Anal Cell Pathol 2019:1038069. https://doi.org/10.1155/2019/1038069
Yu B, Ding YM, Liao XF, Wang CH, Wang B, Chen XY (2019) Overexpression of PARPBP correlates with tumor progression and poor prognosis in HCC. Digest Dis Sci 64(10):2878–2892. https://doi.org/10.1007/s10620-019-05608-4
Zhang M, Wu PB, Li M, Guo YT, Tian T, Liao XC, Tan SY (2021) Inhibition of Notch1 signaling reduces hepatocyte injury in nonalcoholic fatty liver disease via autophagy. Biochem Bioph Res Commun 547:131–138. https://doi.org/10.1016/j.bbrc.2021.02.039
Lu L, Yue S, Jiang LF, Li CY, Zhu Q, Ke M, Lu H, Wang XH, Busuttil RW, Ying QL, Kupiec-Weglinski JW, Ke BB (2018) Myeloid Notch1 deficiency activates the RhoA/ROCK pathway and aggravates hepatocellular damage in mouse ischemic livers. Hepatology 67(3):1041–1055. https://doi.org/10.1002/hep.29593
Li B, Li A, You Z, Xu JC, Zhu S (2020) Epigenetic silencing of CDKN1A and CDKN2B by SNHG1 promotes the cell cycle, migration and epithelial-mesenchymal transition progression of HCC. Cell Death Dis 11(10):823. https://doi.org/10.1038/s41419-020-03031-6
Kadry MO, Abdel-Megeed RM, El-Meliegy E, Abdel-Hamid AHZ (2018) Crosstalk between GSK-3, c-Fos, NF kappa B and TNF-alpha signaling pathways play an ambitious role in chitosan nanoparticles cancer therapy. Toxicol Rep 5:723–727. https://doi.org/10.1016/j.toxrep.2018.06.002
Jia XJ, Guan B, Liao J, Hu XM, Fan Y, Li JH, Zhao HL, Huang QY, Ma ZX, Zhu XF, Fei MX, Lu GD, Nong QQ (2019) Down-regulation of GCLC is involved in microcystin-LR-induced malignant transformation of human liver cells. Toxicology 421:49–58. https://doi.org/10.1016/j.tox.2019.03.010
Xu QS, Fan YH, Loor JJ, Liang YS, Sun XD, Jia HD, Zhao CX, Xu C (2021) Cardamonin reduces acetaminophen-induced acute liver injury in mice via activating autophagy and NFE2L2 signaling. Front Pharmacol 11:601716. https://doi.org/10.3389/fphar.2020.601716
Wojcik KM, Piekarska A, Szymanska B, Jablonowska E (2019) NFE2L2 is associated with NQO1 expression and low stage of hepatic fibrosis in patients with chronic hepatitis C. Adv Clin Exp Med 28(9):1237–1241. https://doi.org/10.17219/acem/105852
Das A, Basu S, Bandyopadhyay D, Mukherjee K, Datta D, Chakraborty S, Jana S, Adak M, Bose S, Chakrabarti S, Swarnakar S, Chakrabarti P, Bhattacharyya SN (2021) Inhibition of extracellular vesicle-associated MMP2 abrogates intercellular hepatic miR-122 transfer to liver macrophages and curtails inflammation. Iscience 24(12):103428. https://doi.org/10.1016/j.isci.2021.103428
Yuan SY, Wei C, Liu GF, Zhang LJ, Li JH, Li LL, Cai SY, Fang L (2022) Sorafenib attenuates liver fibrosis by triggering hepatic stellate cell ferroptosis via HIF-1 alpha/SLC7A11 pathway. Cell Proliferat 55(1):e13158. https://doi.org/10.1111/cpr.13158
Yan JY, Hu BW, Shi WJ, Wang XY, Shen JY, Chen YP, Huang HR, Jin LF (2021) Gli2-regulated activation of hepatic stellate cells and liver fibrosis by TGF-beta signaling. Am J Physiol Gastrointest Liver Physiol 320(5):G720–G728. https://doi.org/10.1152/ajpgi.00310.2020
Jin LF, Huang HR, Ni J, Shen JY, Liu ZP, Li LJ, Fu SM, Yan JY, Hu BW (2021) Shh-Yap signaling controls hepatic ductular reactions in CCl 4 -induced liver injury. Environ Toxicol 36(2):194–203. https://doi.org/10.1002/tox.23025
Park S, Ha YN, Dezhbord M, Lee AR, Park ES, Park YK, Won J, Kim NY, Choo SY, Shin JJ, Ahn CH, Kim KH (2020) Suppression of hepatocyte nuclear factor 4 alpha by long-term infection of hepatitis B virus contributes to tumor cell proliferation. Int J Mol Sci 21(3):948. https://doi.org/10.3390/ijms21030948
Song ZC, Meng LL, He ZX, Huang J, Li F, Feng JJ, Jia ZR, Huang Y, Liu W, Liu AD, Fang HS (2021) LBP protects hepatocyte mitochondrial function via the PPAR-CYP4A2 signaling pathway in a rat sepsis model. Shock 56(6):1066–1079. https://doi.org/10.1097/SHK.0000000000001808
Bi J, Sun K, Wu H, Chen XL, Tang HY, Mao JW (2018) PPAR gamma alleviated hepatocyte steatosis through reducing SOCS3 by inhibiting JAK2/STAT3 pathway. Biochem Bioph Res Commun 498(4):1037–1044. https://doi.org/10.1016/j.bbrc.2018.03.110
Liu YM, Ma JH, Zeng Q, Lv J, Xie XH, Pan YJ, Yu ZJ (2018) MiR-19a affects hepatocyte autophagy via regulating lncRNA NBR2 and AMPK/PPAR in D-GalN/Lipopolysaccharide-stimulated hepatocytes. J Cell Biochem 119(1):358–365. https://doi.org/10.1002/jcb.26188
Feng X, Yu W, Li XD, Zhou FF, Zhang WL, Shen Q, Li JX, Zhang C, Shen PP (2017) Apigenin, a modulator of PPAR gamma, attenuates HFD-induced NAFLD by regulating hepatocyte lipid metabolism and oxidative stress via Nrf2 activation. Biochem Pharmacol 136:136–149. https://doi.org/10.1016/j.bcp.2017.04.014
Zhang QH, Xiang SH, Liu QQ, Gu T, Yao YL, Lu XJ (2018) PPAR gamma antagonizes hypoxia-induced activation of hepatic stellate cell through cross mediating PI3K/AKT and cGMP/PKG signaling. PPAR Res 2018:6970407. https://doi.org/10.1155/2018/6970407
Perri RE, Langer DA, Chatterjee S, Gibbons SJ, Gadgil J, Cao S, Farrugia G, Shah VH (2006) Defects in cGMP-PKG pathway contribute to impaired NO-dependent responses in hepatic stellate cells upon activation. Am J Physiol-Gastr Liver Physiol 290(3):G535–G542. https://doi.org/10.1152/ajpgi.00297.2005
Oliva-Vilarnau N, Vorrink SU, Ingelman-Sundberg M, Lauschke VM (2020) A 3D cell culture model identifies Wnt/beta-catenin mediated inhibition of p53 as a critical step during human hepatocyte regeneration. Adv Sci 7(15):2000248. https://doi.org/10.1002/advs.202000248
Holter MM, Garibay D, Lee SA, Saikia M, McGavigan AK, Ngyuen L, Moore ES, Daugherity E, Cohen P, Kelly K, Weiss RS, Cummings BP (2020) Hepatocyte p53 ablation induces metabolic dysregulation that is corrected by food restriction and vertical sleeve gastrectomy in mice. FASEB J 34(1):1846–1858. https://doi.org/10.1096/fj.201902214R
Zhang JB, Xia Y, Pan W, Zhou DH (2021) Antagonistic effect of nano-selenium on hepatocyte apoptosis induced by DEHP via PI3K/AKT pathway in chicken liver. Ecotox Environ Safe 218:112282. https://doi.org/10.1016/j.ecoenv.2021.112282
Lai PF, Baskaran R, Kuo CH, Day CH, Chen RJ, Ho TJ, Yeh YL, Padma VV, Lai CH, Huang CY (2021) Bioactive dipeptide from potato protein hydrolysate combined with swimming exercise prevents high fat diet induced hepatocyte apoptosis by activating PI3K/Akt in SAMP8 mouse. Mol Biol Rep 48(3):2629–2637. https://doi.org/10.1007/s11033-021-06317-w
Guo JM, Xing HJ, Cai JZ, Zhang HF, Xu SW (2021) H2S exposure-induced oxidative stress promotes LPS-mediated hepatocyte autophagy through the PI3K/AKT/TOR pathway. Ecotox Environ Safe 209:111801. https://doi.org/10.1016/j.ecoenv.2020.111801
Zhang WJ, An R, Li QH, Sun LL, Lai XF, Chen RH, Li DL, Sun SL (2020) Theaflavin TF3 relieves hepatocyte lipid deposition through activating an AMPK signaling pathway by targeting plasma kallikrein. J Agr Food Chem 68(9):2673–2683. https://doi.org/10.1021/acs.jafc.0c00148
Yang YM, Han CY, Kim YJ, Kim SG (2010) AMPK-associated signaling to bridge the gap between fuel metabolism and hepatocyte viability. World J Gastroentero 16(30):3731–3742. https://doi.org/10.3748/wjg.v16.i30.3731
Tsugawa Y, Hiramoto M, Imai T (2019) Estrogen induces estrogen receptor alpha expression and hepatocyte proliferation in late pregnancy. Biochem Bioph Res Commun 511(3):592–596. https://doi.org/10.1016/j.bbrc.2019.02.119
Uebi T, Umeda M, Imai T (2015) Estrogen induces estrogen receptor alpha expression and hepatocyte proliferation in the livers of male mice. Genes Cells 20(3):217–223. https://doi.org/10.1111/gtc.12214
Zhu L, Shi J, Luu TN, Neuman JC, Trefts E, Yu S, Palmisano BT, Wasserman DH, Linton MF, Stafford JM (2018) Hepatocyte estrogen receptor alpha mediates estrogen action to promote reverse cholesterol transport during Western-type diet feeding. Mol Metab 8:106–116. https://doi.org/10.1016/j.molmet.2017.12.012
Song Y, Wu WF, Sheng L, Jiang BJ, Li X, Cai KS (2020) Chrysin ameliorates hepatic steatosis induced by a diet deficient in methionine and choline by inducing the secretion of hepatocyte nuclear factor 4 alpha-dependent very low-density lipoprotein. J Biochem Mol Toxic 34(7):e22497. https://doi.org/10.1002/jbt.22497
Susa N, Ueno S, Furukawa Y, Sugiyama M (1997) Protective effect of deferoxamine on chromium(VI)-induced DNA single-strand breaks, cytotoxicity, and lipid peroxidation in primary cultures of rat hepatocytes. Arch Toxicol 71(6):345–350. https://doi.org/10.1007/s002040050397
Sahu SC, Flynn TJ, Bradlaw JA, Roth WL, Barton CN, Yates JG (2001) Pro-oxidant effects of the flavonoid myricetin on rat hepatocytes in culture. Toxicol Method 11(4):277–283. https://doi.org/10.1080/105172301320254707
Zhang YJ, Xiao F, Liu XM, Liu KH, Zhou XX, Zhong CG (2017) Cr(VI) induces cytotoxicity in vitro through activation of ROS-mediated endoplasmic reticulum stress and mitochondrial dysfunction via the PI3K/Akt signaling pathway. Toxicol In Vitro 41:232–244. https://doi.org/10.1016/j.tiv.2017.03.003
Renu K, Chakraborty R, Myakala H, Koti R, Famurewa AC, Madhyastha H, Vellingiri B, George A, Gopalakrishnan AV (2021) Molecular mechanism of heavy metals (lead, chromium, arsenic, mercury, nickel and cadmium) - induced hepatotoxicity - a review. Chemosphere 271:129735. https://doi.org/10.1016/j.chemosphere.2021.129735
Funding
This study was supported by the Talent Introduction Program of Anhui Science and Technology University (No. DKYJ202003).
Author information
Authors and Affiliations
Contributions
B Yang designed the study. EA Abdel-Moneim reviewed the manuscript and revised the scientific English. XF Li wrote this manuscript and performed the bioinformatics analysis. The authors reviewed this manuscript.
Corresponding author
Ethics declarations
Consent for Publication
All authors agreed to publish this manuscript.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Additional file 1:
All transcripts identified in PRHs of PD-treated and control groups. (XLSX = 3, 032 KB)
Additional file 2:
All genes identified in PRHs of PD-treated and control groups. (XLSX = 1, 318 KB)
Additional file 3:
All DETs in PRHs between PD-treated and control groups. (XLSX = 86 KB)
Additional file 4:
All DEGs in PRHs between PD-treated and control groups. (XLSX = 48 KB)
Additional file 5:
GO analysis for DEGs in PRHs between PD-treated and control groups. (XLSX = 24 KB)
Additional file 6:
KEGG analysis for DEGs in PRHs between PD-treated and control groups. (XLSX = 18 KB)
Additional file 7:
Reactome analysis for DEGs in PRHs between PD-treated and control groups. (XLSX = 16 KB)
Additional file 8:
PANTHER analysis for DEGs in PRHs between PD-treated and control groups. (XLSX = 11 KB)
Additional file 9:
Protein classification for DEGs in PRHs between PD-treated and control groups. (XLSX = 11 KB)
Additional file 10:
GO analysis for hub genes associated with PD-induced hepatotoxicity. (XLSX = 11 KB)
Additional file 11:
KEGG analysis for hub genes associated with PD-induced hepatotoxicity. (XLSX = 11 KB)
Rights and permissions
About this article
Cite this article
Li, X., Abdel-Moneim, AM.E. & Yang, B. Signaling Pathways and Genes Associated with Hexavalent Chromium-Induced Hepatotoxicity. Biol Trace Elem Res 201, 1888–1904 (2023). https://doi.org/10.1007/s12011-022-03291-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12011-022-03291-7