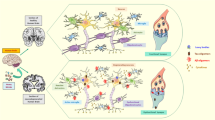
Abstract
Increasing research is illuminating the intricate roles of metal ions in neural development as well as neurological disorders, which may stem from misregulation or dysfunction of epigenetic modifiers. Lead (Pb), cadmium (Cd), aluminum (Al), and arsenic were chosen for critical review because they have become serious public health concerns due to globalization and industrialization. In this review, we will introduce various modes of action of metals and consider the role of two posttranslational modifications: histone acetylation and methylation and how each of them affects gene expression. We then summarize the findings from previous studies on the neurological outcomes and histone alterations in response to the metals on each of the previously described histone modifications mechanisms. Understanding metal-induced histone modifications changes could provide better insight on the mechanism through which neurotoxicity occurs, to propose and validate these modifications as possible biomarkers for early identification of neurological damage, and can help model targeted therapies for the diseases of the brain.

Similar content being viewed by others
References
Karri V, Schuhmacher M, Kumar V (2016) Heavy metals (Pb, Cd, As and MeHg) as risk factors for cognitive dysfunction: a general review of metal mixture mechanism in brain. Environ Toxicol Pharmacol 48:203–213
Colomina MT, Peris-Sampedro F (2017) Aluminum and Alzheimer's disease. Adv Neurobiol 18:183–197
Hesdorffer DC (2016) Comorbidity between neurological illness and psychiatric disorders. CNS Spectr 21(3):230–238
Wang T, Zhang J and Xu Y, Epigenetic basis of lead-induced neurological disorders. Int J Environ Res Public Health, 2020. 17(13).
Bihaqi SW et al (2011) Infant exposure to lead (Pb) and epigenetic modifications in the aging primate brain: implications for Alzheimer's disease. J Alzheimers Dis 27(4):819–833
Chatterjee D et al (2018) Role of microRNAs in senescence and its contribution to peripheral neuropathy in the arsenic exposed population of West Bengal. India. Environ Pollut 233:596–603
Raciti M, Ceccatelli S (2018) Epigenetic mechanisms in developmental neurotoxicity. Neurotoxicol Teratol 66:94–101
Du X et al (2018) Cortex and hippocampus DNA epigenetic response to a long-term arsenic exposure via drinking water. Environ Pollut 234:590–600
Guida N et al (2016) Methylmercury upregulates RE-1 silencing transcription factor (REST) in SH-SY5Y cells and mouse cerebellum. Neurotoxicology 52:89–97
Shi L, Wen H, Shi X (2017) The histone variant H3.3 in transcriptional regulation and human disease. J Mol Biol 429(13):1934–1945
Ramazi S, Allahverdi A and Zahiri J, Evaluation of post-translational modifications in histone proteins: a review on histone modification defects in developmental and neurological disorders. J Biosci, 2020. 45.
Bonnaud EM, Suberbielle E, Malnou CE (2016) Histone acetylation in neuronal (dys)function. Biomol Concepts 7(2):103–116
Angarica VE, Del SA (2017) Bioinformatics tools for genome-wide epigenetic research. Adv Exp Med Biol 978:489–512
Yao B et al (2016) Epigenetic mechanisms in neurogenesis. Nat Rev Neurosci 17(9):537–549
Barnes CE, English DM, Cowley SM (2019) Acetylation & Co: an expanding repertoire of histone acylations regulates chromatin and transcription. Essays Biochem 63(1):97–107
Wapenaar H, Dekker FJ (2016) Histone acetyltransferases: challenges in targeting bi-substrate enzymes. Clin Epigenetics 8:59
Sheikh BN (2014) Crafting the brain - role of histone acetyltransferases in neural development and disease. Cell Tissue Res 356(3):553–573
Seto E, Yoshida M (2014) Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb Perspect Biol 6(4):a018713
Vandebroek A, Yasui M (2020) Regulation of AQP4 in the central nervous system. Int J Mol Sci:21(5)
Demyanenko S et al (2019) capital ES, Cyrilliclass II histone deacetylases in the post-stroke recovery period-expression, cellular, and subcellular localization-promising targets for neuroprotection. J Cell Biochem 120(12):19590–19609
Anamika et al (2019) Mitochondrial SIRT3 and neurodegenerative brain disorders. J Chem Neuroanat 95:43–53
Cao J et al (2019) HDAC11 regulates type I interferon signaling through defatty-acylation of SHMT2. Proc Natl Acad Sci U S A 116(12):5487–5492
He K, Cao X, Deng X (2021) Histone methylation in epigenetic regulation and temperature responses. Curr Opin Plant Biol 61:102001
Pattaroni C, Jacob C (2013) Histone methylation in the nervous system: functions and dysfunctions. Mol Neurobiol 47(2):740–756
Sterling J, et al., Histone lysine demethylases and their functions in cancer. Int J Cancer, 2020.
Collins BE et al (2019) Histone H3 lysine K4 methylation and its role in learning and memory. Epigenetics Chromatin 12(1):7
Guan JS et al (2009) HDAC2 negatively regulates memory formation and synaptic plasticity. Nature 459(7243):55–60
Wang W, et al., Epigenetic treatment of behavioral and physiological deficits in a tauopathy mouse model. Aging Cell, 2021: p. e13456.
Igarashi S et al (2003) Inducible PC12 cell model of Huntington's disease shows toxicity and decreased histone acetylation. Neuroreport 14(4):565–568
Lee MJ et al (2019) Calebin-A induced death of malignant peripheral nerve sheath tumor cells by activation of histone acetyltransferase. Phytomedicine 57:377–384
Subbanna S, et al., Ethanol exposure induces neonatal neurodegeneration by enhancing CB1R Exon1 histone H4K8 acetylation and up-regulating CB1R function causing neurobehavioral abnormalities in adult mice. Int J Neuropsychopharmacol, 2014. 18(5).
Wang S, et al., Selenium-alleviated testicular toxicity by modulating inflammation, heat shock response, and autophagy under oxidative stress in lead-treated chickens. Biol Trace Elem Res, 2021.
Li N et al (2021) Protective effects of folic acid on oxidative damage of rat spleen induced by lead acetate. Ecotoxicol Environ Saf 211:111917
Gao K et al (2020) The role of endoplasmic reticulum stress in lead (Pb)-induced mitophagy of HEK293 cells. Toxicol Ind Health 36(12):1002–1009
Alsadany MA et al (2013) Histone deacetylases enzyme, copper, and IL-8 levels in patients with Alzheimer's disease. Am J Alzheimers Dis Other Demen 28(1):54–61
Gu X et al (2019) Interplay of miR-137 and EZH2 contributes to the genome-wide redistribution of H3K27me3 underlying the Pb-induced memory impairment. Cell Death Dis 10(9):671
Eid A et al (2018) Histone acetylation maps in aged mice developmentally exposed to lead: epigenetic drift and Alzheimer-related genes. Epigenomics 10(5):573–583
Wu Y et al (2018) Regulatory roles of histone deacetylases 1 and 2 in Pb-induced neurotoxicity. Toxicol Sci 162(2):688–701
Wang Y et al (2019) Latent role of in vitro Pb exposure in blocking Abeta clearance and triggering epigenetic modifications. Environ Toxicol Pharmacol 66:14–23
Luo M et al (2014) Epigenetic histone modification regulates developmental lead exposure induced hyperactivity in rats. Toxicol Lett 225(1):78–85
Gu X et al (2020) Nuclear accumulation of histone deacetylase 4 (HDAC4) by PP1-mediated dephosphorylation exerts neurotoxicity in Pb-exposed neural cells. Neurotoxicology 81:395–405
Lin LF et al (2021) Low dose lead exposure induces alterations on heterochromatin hallmarks persisting through SH-SY5Y cell differentiation. Chemosphere 264(Pt 1):128486
Niu Y et al (2015) Oxidative stress alters global histone modification and DNA methylation. Free Radic Biol Med 82:22–28
Dave N et al (2021) Identification of retinoblastoma binding protein 7 (Rbbp7) as a mediator against tau acetylation and subsequent neuronal loss in Alzheimer's disease and related tauopathies. Acta Neuropathol 142(2):279–294
Moyano P et al (2018) Cadmium induced ROS alters M1 and M3 receptors, leading to SN56 cholinergic neuronal loss, through AChE variants disruption. Toxicology 394:54–62
Monaco A et al (2017) Neurodegeneration in zebrafish embryos and adults after cadmium exposure. Eur J Histochem 61(4):2833
Lamtai M et al (2021) Melatonin ameliorates cadmium-induced affective and cognitive impairments and hippocampal oxidative stress in Rat. Biol Trace Elem Res 199(4):1445–1455
Zhou R et al (2020) Combined exposure of lead and cadmium leads to the aggravated neurotoxicity through regulating the expression of histone deacetylase 2. Chemosphere 252:126589
Jacob S, Sumathi T (2019) Extenuation of in utero toxic effects of MeHg in the developing neurons by Fisetin via modulating the expression of synaptic transmission and plasticity regulators in hippocampus of the rat offspring. Chem Biol Interact 305:3–10
Onishchenko N et al (2008) Long-lasting depression-like behavior and epigenetic changes of BDNF gene expression induced by perinatal exposure to methylmercury. J Neurochem 106(3):1378–1387
Go S, et al., DNA methyltransferase- and histone deacetylase-mediated epigenetic alterations induced by low-level methylmercury exposure disrupt neuronal development. Arch Toxicol, 2021.
Guida N et al (2017) p38/Sp1/Sp4/HDAC4/BDNF axis is a novel molecular pathway of the neurotoxic effect of the methylmercury. Front Neurosci 11:8
Go S et al (2018) Methylmercury causes epigenetic suppression of the tyrosine hydroxylase gene in an in vitro neuronal differentiation model. Biochem Biophys Res Commun 502(4):435–441
Goetzl EJ et al (2015) Low neural exosomal levels of cellular survival factors in Alzheimer's disease. Ann Clin Transl Neurol 2(7):769–773
Guida N et al (2018) The miR206-JunD circuit mediates the neurotoxic effect of methylmercury in cortical neurons. Toxicol Sci 163(2):569–578
Pan B et al (2020) Relationship between occupational aluminium exposure and histone lysine modification through methylation. J Trace Elem Med Biol 61:126551
Qiu HY et al (2016) Association between H3K4me3/BDNF and the cognitive function of workers occupationally exposed to aluminum. Zhonghua Lao Dong Wei Sheng Zhi Ye Bing Za Zhi 34(12):900–904
Li J, et al., Protective effects of low-intensity pulsed ultrasound on aluminum overload-induced cerebral damage through epigenetic regulation of brain-derived neurotrophic factor expression. Biosci Rep, 2019. 39(1).
Li H et al (2020) Aluminium-induced synaptic plasticity injury via the PHF8-H3K9me2-BDNF signalling pathway. Chemosphere 244:125445
Li Z et al (2016) The pilot study on the expression of PHF8, H3K9me2, BDNF and LTP in the hippocampus of rats exposed to aluminum. Zhonghua Lao Dong Wei Sheng Zhi Ye Bing Za Zhi 34(1):18–22
Wang F et al (2019) Role of MLL in the modification of H3K4me3 in aluminium-induced cognitive dysfunction. Chemosphere 232:121–129
Yu X, et al., Chitotriosidase attenuates brain inflammation via HDAC3/NF-κB pathway in D-galactose and aluminum-induced rat model with cognitive impairments. Neuroscience Research, 2021.
Hasanvand M et al (2020) Dose-response meta-analysis of arsenic exposure in drinking water and intelligence quotient. Journal of Environmental Health Science and Engineering 18(2):1691–1697
Tyler CR et al (2015) Developmental exposure to 50 parts-per-billion arsenic influences histone modifications and associated epigenetic machinery in a region- and sex-specific manner in the adult mouse brain. Toxicol Appl Pharmacol 288(1):40–51
Solomon ER, Caldwell KK, Allan AM (2020) Developmental arsenic exposure is associated with sex differences in the epigenetic regulation of stress genes in the adult mouse frontal cortex. Toxicol Appl Pharmacol 391:114920
Chervona Y et al (2012) Associations between arsenic exposure and global posttranslational histone modifications among adults in Bangladesh. Cancer Epidemiol Biomarkers Prev 21(12):2252–2260
Cronican AA et al (2013) Genome-wide alteration of histone H3K9 acetylation pattern in mouse offspring prenatally exposed to arsenic. PLoS One 8(2):e53478
Tyler C et al (2018) Sex-dependent effects of the histone deacetylase inhibitor, sodium valproate, on reversal learning after developmental arsenic exposure. Front Genet 9:200
Kim HY et al (2016) Differential epigenetic effects of chlorpyrifos and arsenic in proliferating and differentiating human neural progenitor cells. Reprod Toxicol 65:212–223
Soh M et al (2012) Increased neuron specific enolase expression by urothelial cells exposed to or malignantly transformed by exposure to Cd(2)(+) or As(3)(+). Toxicol Lett 212(1):66–74
Karri V, Schuhmacher MP, Kumar V (2020) A systems toxicology approach to compare the heavy metal mixtures (Pb, As, MeHg) impact in neurodegenerative diseases. Food Chem Toxicol 139:111257
Inhalable metal-rich Air particles and histone H3K4 dimethylation and H3K9 acetylation in a cross-sectional study of steel workers.
Ding R et al (2017) Dose- and time- effect responses of DNA methylation and histone H3K9 acetylation changes induced by traffic-related air pollution. Sci Rep 7:43737
Morgan M, Shilatifard A (2020) Reevaluating the roles of histone-modifying enzymes and their associated chromatin modifications in transcriptional regulation. Nat Genet 52(12):1271–1281
Ijomone OM et al (2020) Epigenetic influence of environmentally neurotoxic metals. Neurotoxicology 81:51–65
Wang B et al (2012) Cadmium and its epigenetic effects. Curr Med Chem 19(16):2611–2620
Culbreth M, Aschner M (2019) Methylmercury epigenetics. Toxics:7(4)
Liang R (2018) Cross talk between aluminum and genetic susceptibility and epigenetic modification in Alzheimer's disease. Adv Exp Med Biol 1091:173–191
Garza-Lombo C et al (2019) Arsenic-induced neurotoxicity: a mechanistic appraisal. J Biol Inorg Chem 24(8):1305–1316
Hashimoto H, Vertino PM, Cheng X (2010) Molecular coupling of DNA methylation and histone methylation. Epigenomics 2(5):657–669
Author information
Authors and Affiliations
Contributions
All authors contributed to the preparation of this review.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wu, Y., Wang, R., Liu, R. et al. The Roles of Histone Modifications in Metal-Induced Neurological Disorders. Biol Trace Elem Res 201, 31–40 (2023). https://doi.org/10.1007/s12011-022-03134-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12011-022-03134-5