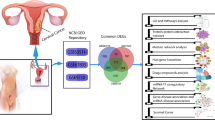
Abstract
Differently expressed genes (DEGs) across cervical (CC), endometrial (EC), and vulvar carcinoma (VC) may serve as potential biomarkers for these progressive tumor conditions. In this study, DEGs of cervical (CC), endometrial (EC), and vulvar carcinoma (VC) were identified by microarray analysis. The interaction network between the identified 124 DEGs was constructed and analyzed to identify the hub genes and genes with high stress centrality. DEGs, namely, CDK1 and MMP9, were found to show highest degree and highest stress centrality respectively from the gene interaction network of 124 nodes and 1171 edges. DEG CDK1 is found to be overlapping in both cervical and endometrial carcinomic conditions while DEG MMP9 is found in vulvar carcinomic condition. Further, as it is studied that many phytochemicals play an important role as medicinal drugs, we have identified phytochemicals from few widely available medicinal plants and performed comprehensive computational study to identify a multi-targeted phytochemical against the identified DEGs, which are crucially responsible for the progression of these carcinomic conditions. Virtual screening of the phytochemicals against the target DEG protein structures with PDB IDs 4Y72 and 1GKC resulted in identifying the multi-targeted phytochemical against both the proteins. The molecular docking and dynamics simulation studies reveal that luteolin can act as a multi-targeted agent. Thus, the interactional and structural insights of luteolin toward the DEG proteins signify that it can be further explored as a multi-targeted agent against the cervical, endometrial, and vulvar carcinoma.










Similar content being viewed by others
Data Availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Code Availability
Not applicable.
References
Pappa, K. I., Polyzos, A., Jacob-Hirsch, J., et al. (2015). Profiling of discrete gynecological cancers reveals novel transcriptional modules and common features shared by other cancer types and embryonic stem cells. PloS One, 10, e0142229. https://doi.org/10.1371/journal.pone.0142229
Zhang, S., Xu, H., Zhang, L., et al. (2020). Cervical cancer: Epidemiology, risk factors and screening. Chinese Journal of Cancer Research, 32(6), 720–728. https://doi.org/10.21147/J.ISSN.1000-9604.2020.06.05
Balasubramaniam, S. D., Balakrishnan, V., Oon, C. E., & Kaur, G. (2019). Key molecular events in cervical cancer development. Medicina, 55(7), 384. https://doi.org/10.3390/MEDICINA55070384
van Weelden, W. J., Massuger, L. F. A. G., ENITEC, et al. (2019). Anti-estrogen treatment in endometrial cancer: A systematic review. Frontiers in Oncology, 0, 359. https://doi.org/10.3389/FONC.2019.00359
Samarnthai, N., Hall, K., & Yeh, I.-T. (2010). Molecular profiling of endometrial malignancies. Obstetrics and Gynecology International, 2010, 1–16. https://doi.org/10.1155/2010/162363
Rogers, L. J., & Cuello, M. A. (2018). Cancer of the vulva. International Journal of Gynecology & Obstetrics, 143, 4–13. https://doi.org/10.1002/IJGO.12609
Mantovani, G., Fragomeni, S. M., Inzani, F., et al. (2020). Molecular pathways in vulvar squamous cell carcinoma: Implications for target therapeutic strategies. Journal of Cancer Research and Clinical Oncology, 146(7), 1647–1658. https://doi.org/10.1007/S00432-020-03226-6
Slack, J. M. W. (2012). Essential developmental biology. Wiley.
Winder, D. M., Chattopadhyay, A., Muralidhar, B., et al. (2011). Overexpression of the oncostatin M receptor in cervical squamous cell carcinoma cells is associated with a pro-angiogenic phenotype and increased cell motility and invasiveness. The Journal of Pathology, 225, 448–462.
Zhai, Y., Kuick, R., Nan, B., et al. (2007). Gene expression analysis of preinvasive and invasive cervical squamous cell carcinomas identifies HOXC10 as a key mediator of invasion. Cancer Research, 67, 10163–10172.
Pappa, K. I., Jacob-Hirsch, J., Vlachos, G. D., et al. (2011). Expression profiling of vulvar carcinoma: Clues for deranged extracellular matrix remodeling and effects on multiple signaling pathways combined with discrete patient subsets. Translational Oncology, 4, 301–IN6.
Wu, X., Chen, L., & Wang, X. (2014). Network biomarkers, interaction networks and dynamical network biomarkers in respiratory diseases. Clinical and Translational Medicine, 3, 1–7.
Guo, L., Du, Y., & Wang, J. (2015). Network analysis reveals a stress-affected common gene module among seven stress-related diseases/systems which provides potential targets for mechanism research. Scientific Reports, 5, 1–6.
Suman, S., & Mishra, A. (2018). An interaction network driven approach for identifying biomarkers for progressing cervical intraepithelial neoplasia. Scientific Reports, 8, 1–11.
Kumbhar, S. T., Patil, S. P., & Une, H. D. (2018). Phytochemical analysis of Canna indica L. roots and rhizomes extract. Biochemistry and Biophysics Reports, 16, 50–55.
Sarje, S. K., Ingole, K., Angad, S., et al. (2019). A pharmacognostic and pharmacological review on Canna indica Linn. International Journal of Research in Pharmacy and Chemistry, 9, 61–77.
Anjana, G. V., Priya, D., Srimathi, R., & KB, S. H. A. N. T. H. A. (2018). A review on medical advantages and chemical constituents of Carica papaya Linn. Asian Journal of Pharmaceutical and Clinical Research, 11, 53–57.
Yogiraj, V., Goyal, P. K., Chauhan, C. S., et al. (2014). Carica papaya Linn: An overview. International Journal of Herbal Medicine, 2, 01–08.
Igwe, O. U. (2015). Chemical constituents of the leaf essential oil of Carica papaya from South East Nigeria and its antimicrobial activity. IJRPC, 5, 77–83.
Oche, O., Rosemary, A., John, O., et al. (2017). Chemical constituents and nutrient composition of Carica papaya and Vernonia amygdalina leaf extracts. Journal of Complementary and Alternative Medical Research, 1–8.
Ved, A., Arsi, T., Prakash, O., & Gupta, A. (2018). A review on phytochemistry and pharmacological activity of Lantana camara Linn. International Journal of Pharmaceutical Sciences and Research, 9, 37–43.
Jurenka, J. (2008). Therapeutic applications of pomegranate (Punica granatum L.): A review. Alternative Medicine Review, 13.
Wu, S., & Tian, L. (2017). Diverse phytochemicals and bioactivities in the ancient fruit and modern functional food pomegranate (punica granatum). Molecules, 22.
Zhang, W., Gao, L., Wang, C., et al. (2020). Combining bioinformatics and experiments to identify and verify key genes with prognostic values in endometrial carcinoma. Journal of Cancer, 11, 716–732. https://doi.org/10.7150/jca.35854
Liu, L., Lin, J., & He, H. (2019). Identification of potential crucial genes associated with the pathogenesis and prognosis of endometrial cancer. Frontiers in Genetics, 10, 373. https://doi.org/10.3389/fgene.2019.00373
Shi, S., Tan, Q., Feng, F., et al. (2020). Identification of core genes in the progression of endometrial cancer and cancer cell-derived exosomes by an integrative analysis. Scientific Reports, 10, 1–14. https://doi.org/10.1038/s41598-020-66872-3
Bioconductor. (2008). In: Encyclopedia of genetics, genomics, proteomics and informatics (p. 211). Springer.
Benjamini, Y., & Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B (Methodological), 57, 289–300. https://doi.org/10.1111/j.2517-6161.1995.tb02031.x
Szklarczyk, D., Gable, A. L., Lyon, D., et al. (2019). STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Research, 47, D607–D613. https://doi.org/10.1093/nar/gky1131
Shannon, P. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Research, 13, 2498–2504. https://doi.org/10.1101/gr.1239303
Friesner, R. A., Banks, J. L., Murphy, R. B., et al. (2004). Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. Journal of Medicinal Chemistry, 47, 1739–1749.
O’Boyle, N. M., Banck, M., James, C. A., et al. (2011). Open Babel: An open chemical toolbox. Journal of cheminformatics, 3, 1–14.
Shelley, J. C., Cholleti, A., Frye, L. L., et al. (2007). Epik: A software program for pK a prediction and protonation state generation for drug-like molecules. Journal of Computer-Aided Molecular Design, 21, 681–691.
Van Der Spoel, D., Lindahl, E., Hess, B., et al. (2005). GROMACS: Fast, flexible, and free. Journal of Computational Chemistry, 26, 1701–1718.
Huang, J., Rauscher, S., Nawrocki, G., et al. (2017). CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nature Methods, 14, 71–73.
Zoete, V., Cuendet, M. A., Grosdidier, A., & Michielin, O. (2011). SwissParam: A fast force field generation tool for small organic molecules. Journal of Computational Chemistry, 32, 2359–2368.
Darden, T., York, D., & Pedersen, L. (1993). Particle mesh Ewald: An N· log (N) method for Ewald sums in large systems. The Journal of Chemical Physics, 98, 10089–10092.
Maurya, A. K., Mulpuru, V., & Mishra, N. (2020). Discovery of novel coumarin analogs against the α-glucosidase protein target of diabetes mellitus: Pharmacophore-based QSAR, docking, and molecular dynamics simulation studies. ACS omega.
Kumari, R., Kumar, R., OSDD, C., & Lynn, A. (2014). g_mmpbsa—A GROMACS tool for high-throughput MM-PBSA calculations. Journal of Chemical Information and Modeling, 54, 1951–1962.
Zhang, M., Zhang, L., Hei, R., et al. (2021). CDK inhibitors in cancer therapy, an overview of recent development. American Journal of Cancer Research, 11, 1913.
Huang, H. (2018). Matrix metalloproteinase-9 (MMP-9) as a cancer biomarker and MMP-9 biosensors: Recent advances. Sensors (Basel, Switzerland), 18. https://doi.org/10.3390/S18103249
Fields, G. B. (2019). The rebirth of matrix metalloproteinase inhibitors: Moving beyond the dogma. Cells, 8. https://doi.org/10.3390/CELLS8090984
Brown, N. R., Korolchuk, S., Martin, M. P., et al. (2015). CDK1 structures reveal conserved and unique features of the essential cell cycle CDK. Nature Communications, 6, 6769. https://doi.org/10.1038/ncomms7769
Zheng, H.-P., Huang, Z.-G., He, R.-Q., et al. (2019). Integrated assessment of CDK1 upregulation in thyroid cancer. American Journal of Translational Research, 11, 7233–7254.
Prevo, R., Pirovano, G., Puliyadi, R., et al. (2018). CDK1 inhibition sensitizes normal cells to DNA damage in a cell cycle dependent manner. Cell cycle (Georgetown, Tex), 17, 1513–1523. https://doi.org/10.1080/15384101.2018.1491236
Mondal, S., Adhikari, N., Banerjee, S., et al. (2020). Matrix metalloproteinase-9 (MMP-9) and its inhibitors in cancer: A minireview. European Journal of Medicinal Chemistry, 194, 112260. https://doi.org/10.1016/j.ejmech.2020.112260
Lin, Y., Shi, R., Wang, X., & Shen, H.-M. (2008). Luteolin, a flavonoid with potential for cancer prevention and therapy. Current Cancer Drug Targets, 8, 634–646. https://doi.org/10.2174/156800908786241050
Imran, M., Rauf, A., Abu-Izneid, T., et al. (2019). Luteolin, a flavonoid, as an anticancer agent: A review. Biomedicine & Pharmacotherapy, 112, 108612. https://doi.org/10.1016/J.BIOPHA.2019.108612
Tuorkey, M. J. (2016). Molecular targets of luteolin in cancer. European Journal of Cancer Prevention, 25, 65–76. https://doi.org/10.1097/CEJ.0000000000000128
David, C. C., & Jacobs, D. J. (2014). Principal component analysis: A method for determining the essential dynamics of proteins. Methods in Molecular Biology, 1084, 193–226. https://doi.org/10.1007/978-1-62703-658-0_11
Acknowledgements
The research work was carried out in the Laboratory of Chemistry, Department of Applied Sciences, Indian Institute of Information Technology Allahabad, Prayagraj, India. The molecular dynamic simulation results reported in this work were performed on the Central Computing Facility of IIITA, Prayagraj.
Author information
Authors and Affiliations
Contributions
A.M. and N.M. conceived the study, A.M. took part in the network analysis and the docking studies, and V.M. took part in the molecular dynamics simulation studies. A.M. and V.M. wrote the main manuscript and prepared the figures. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mishra, A., Mulpuru, V. & Mishra, N. An Interaction Network Driven Approach for Identifying Cervical, Endometrial, Vulvar Carcinomic Biomarkers and Their Multi-targeted Inhibitory Agents from Few Widely Available Medicinal Plants. Appl Biochem Biotechnol 195, 6893–6912 (2023). https://doi.org/10.1007/s12010-023-04441-w
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-023-04441-w