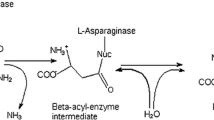
Abstract
S-adenosyl-L-methionine (SAM) is the active form of methionine, which participates in various metabolic reactions and plays a vital role. It is mainly used as a precursor by three key metabolic pathways: trans-methylation, trans-sulfuration, and trans-aminopropylation. Methionine adenosyltransferase (MAT) is the only enzyme to produce SAM from methionine and ATP. However, there is no efficient and accurate method for high-throughput detection of SAM, which is the major obstacles of directed evolution campaigns for MAT. Herein, we established a colorimetric method for directed evolution of MAT based on detecting SAM by using glycine oxidase and glycine/sarcosine N-methyltransferase enzyme. Screening of MAT libraries revealed variant I303V/Q22R with 2.13-fold improved activity towards SAM in comparison to the wild type. Molecular dynamic simulation indicates that the loops more flexible and more conducive to SAM release.







Similar content being viewed by others
Data Availability
The data presented in this study are available on request from the corresponding author.
References
Giulidori, P., Galli Kienle, M., Catto, E., & Stramentinoli, G. (1984). Transmethylation, transsulfuration, and aminopropylation reactions of S-adenosyl-L-methionine in vivo. The Journal of Biological Chemistry, 259, 4205–4211.
Lu, S. C. (2000). S-Adenosylmethionine. The International Journal of Biochemistry & Cell Biology, 32, 391–395.
Liu, Y., Chen, B., Wang, Z., Liu, L., & Tan, T. (2016). Functional characterization of a thermostable methionine adenosyltransferase from Thermus thermophilus HB27. Frontiers of Chemical Science and Engineering, 10, 238–244.
Markham, G. D., Hafner, E. W., Tabor, C. W., & Tabor, H. (1980). S-adenosylmethionine synthetase from Escherichia coli. Journal of Biological Chemistry, 255, 9082–9092.
McQueney, M. S., & Markham, G. D. (1995). Investigation of monovalent cation activation of S-adenosylmethionine synthetase using mutagenesis and uranyl inhibition. Journal of Biological Chemistry, 270, 18277–18284.
Chiang, P. K., & Cantoni, G. L. (1977). Activation of methionine for transmethylation. Purification of the S adenosylmethione synthetase of bakers’ yeast and its separation into two forms. The Journal of Biological Chemistry, 252, 4506–4513.
He, J., Deng, J., Zheng, Y., & Gu, J. (2006). A synergistic effect on the production of S-adenosyl-l-methionine in Pichia pastoris by knocking in of S-adenosyl-l-methionine synthase and knocking out of cystathionine-β synthase. Journal of Biotechnology, 126, 519–527.
Hu, H., et al. (2009). Optimization of l-methionine feeding strategy for improving S-adenosyl-l-methionine production by methionine adenosyltransferase overexpressed Pichia pastoris. Applied Microbiology and Biotechnology, 83, 1105–1114.
Yin, C., Zheng, T. & Chang, X. (2017) Biosynthesis of S-adenosylmethionine by magnetically immobilized Escherichia coli cells highly expressing a methionine adenosyltransferase variant. Molecules 22
Matos, J. R., & Wong, C. H. (1987). S-adenosylmethionine: Stability and stabilization. Bioorganic Chemistry, 15, 71–80.
Wang, X., et al. (2019). Semi-rationally engineered variants of S-adenosylmethionine synthetase from Escherichia coli with reduced product inhibition and improved catalytic activity. Enzyme and Microbial Technology, 129, 109355.
Dippe, M., et al. (2015). Rationally engineered variants of S-adenosylmethionine (SAM) synthase: Reduced product inhibition and synthesis of artificial cofactor homologues. Chemical Communications, 51, 3637–3640.
Weiß, M. S., Pavlidis, I. V., Vickers, C., Hohne, M., & Bornscheuer, U. T. (2014). Glycine oxidase based high-throughput solid-phase assay for substrate profiling and directed evolution of (R)- and (S)-selective amine transaminases. Analytical Chemistry, 86, 11847–11853.
Ogawa, H., Gomi, T., Takusagawa, F., & Fujioka, M. (1998). Structure, function and physiological role of glycine N- methyltransferase. The International Journal of Biochemistry & Cell Biology, 30, 13–26.
Waditee, R., et al. (2003). Isolation and functional characterization of N-methyltransferases that catalyze betaine synthesis from glycine in a halotolerant photosynthetic organism Aphanothece halophytica. Journal of Biological Chemistry, 278, 4932–4942.
Shi, Y., et al. (2019). Atp mimics ph-dependent dual peroxidase-catalase activities driving H2O2 decomposition. CCS Chemistry, 1, 373–383.
Takata, Y., et al. (2003). Catalytic mechanism of glycine N-methyltransferase. Biochemistry, 42, 8394–8402.
Hu, H., et al. (2009). DNA shuffling of methionine adenosyltransferase gene leads to improved S-adenosyl-l-methionine production in Pichia pastoris. Journal of Biotechnology, 141, 97–103.
Kamarthapu, V., Rao, K. V., Srinivas, P. N. B. S., Reddy, G. B., & Reddy, V. D. (2008). Structural and kinetic properties of Bacillus subtilis S-adenosylmethionine synthetase expressed in Escherichia coli. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics, 1784, 1949–1958.
Han, G., Hu, X., & Wang, X. (2016). Overexpression of methionine adenosyltransferase in Corynebacterium glutamicum for production of S-adenosyl-l-methionine. Biotechnology and Applied Biochemistry, 63, 679–689.
Park, J., Tai, J., Roessner, C. A., & Scott, A. I. (1995). Overcoming product inhibition of S-Adenosyl-L-methionine (SAM) synthetase: Preparation of SAM on the 30 mM scale. Bioorganic & Medicinal Chemistry Letters, 5, 2203–2206.
Gilliland, G. L., Markham, G. D., & Davies, D. R. (1983). S-adenosylmethionine synthetase from Escherichia coli. Crystallization and preliminary X-ray diffraction studies. The Journal of Biological Chemistry, 258, 6963–6964.
Schalk-Hihi, C., & Markham, G. D. (1999). The conformations of a substrate and a product bound to the active site of S-adenosylmethionine synthetase. Biochemistry, 38, 2542–2550.
Park, J., Tai, J., Roessner, C. A., & Scott, A. I. (1996). Enzymatic synthesis of S-adenosyl-L-methionine on the preparative scale. Bioorganic & Medicinal Chemistry, 4, 2179–2185.
Fu, Z., Hu, Y., Markham, G. D., & Takusagawa, F. (1996). Flexible loop in the structure of s-adenosylmethionine synthetase crystallized in the tetragonal modification. Journal of Biomolecular Structure and Dynamics, 13, 727–739.
Taylor, J. C., Takusagawa, F., & Markham, G. D. (2002). The active site loop of S-adenosylmethionine synthetase modulates catalytic efficiency. Biochemistry, 41, 9358–9369.
Funding
This study was funded by the National Key Research and Development Program of China (grant number 2021YFC2101000). This study was also funded by the National Natural Science Foundation of China (grant numbers 52073022, 21978017).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by Chennqi Cao and Kaili Nie. The first draft of the manuscript was written by Chennqi Cao, Haijun Xu, and Luo Liu, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cao, C., Nie, K., Xu, H. et al. High-Throughput Screening and Directed Evolution of Methionine Adenosyltransferase from Escherichia coli. Appl Biochem Biotechnol 195, 4053–4066 (2023). https://doi.org/10.1007/s12010-023-04314-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-023-04314-2