Abstract
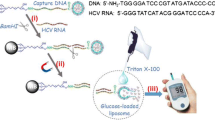
Exosome plays a crucial role in regulating intercellular communication during atherosclerosis development. However, sensitive and portable exosome detection remains a huge challenge. Herein, a personal glucose meter (PGM)-based exosomes detection approach has been proposed that allows detection of exosomes with a high sensitivity and reproducibility. In this method, a catch probe, which is composed of CD63 aptamer and blocker sequence, is utilized for the specific identification of exosomes. The blocker sequence binds with H probe to initiate the Exo-III-assisted signal recycles to generate numerous DNAzyme sequences. Under the assistance of the substrate, DNAzyme forms its active secondary structure to generate gap site in substrate, releasing a linker to conjugate sucrase to streptavidin magnetic beads (SMBs). After removing unbound sucrase, the SMB-linker-sucrase complex is used to catalyze sucrose to glucose, which can be read by PGMs. Based on this, the method exhibits a wide detection range and a low limit of detection, holding a promising prospect for the analysis of exosomes and screening atherosclerosis.
Graphical abstract





Similar content being viewed by others
Data Availability
All data generated and analyzed during this study are included in this article.
References
Wang, C., Li, Z., Liu, Y., & Yuan, L. (2021). Exosomes in atherosclerosis: Performers, bystanders, biomarkers, and therapeutic targets. Theranostics, 11, 3996–4010.
Wang, Y., Xie, Y., Zhang, A., Wang, M., Fang, Z., & Zhang, J. (2019). Exosomes: An emerging factor in atherosclerosis. Biomedicine & Pharmacotherapy, 115, 108951.
Gao, W., Liu, H., Yuan, J., Wu, C., Huang, D., Ma, Y., Zhu, J., Ma, L., Guo, J., Shi, H., Zou, Y., & Ge, J. (2016). Exosomes derived from mature dendritic cells increase endothelial inflammation and atherosclerosis via membrane TNF-alpha mediated NF-kappaB pathway. Journal of Cellular and Molecular Medicine, 20, 2318–2327.
Henning, R. J. (2021). Cardiovascular exosomes and microRNAs in cardiovascular physiology and pathophysiology. Journal of Cardiovascular Translational Research, 14, 195–212.
Ling, H., Guo, Z., Tan, L., Cao, Q., & Song, C. (2021). Stem cell-derived exosomes: Role in the pathogenesis and treatment of atherosclerosis. International Journal of Biochemistry & Cell Biology, 130, 105884.
Zheng, D., Huo, M., Li, B., Wang, W., Piao, H., Wang, Y., Zhu, Z., Li, D., Wang, T., & Liu, K. (2020). The role of exosomes and exosomal microRNA in cardiovascular disease. Front Cell Dev Biol, 8, 616161.
Lin, B., Yang, J., Song, Y., Dang, G., & Feng, J. (2021). Exosomes and atherogenesis. Front Cardiovasc Med, 8, 738031.
Becker, A., Thakur, B. K., Weiss, J. M., Kim, H. S., Peinado, H., & Lyden, D. (2016). Extracellular vesicles in cancer: Cell-to-cell mediators of metastasis. Cancer Cell, 30, 836–848.
Maacha, S., Bhat, A. A., Jimenez, L., Raza, A., Haris, M., Uddin, S., & Grivel, J. C. (2019). Extracellular vesicles-mediated intercellular communication: Roles in the tumor microenvironment and anti-cancer drug resistance. Molecular Cancer, 18, 55.
Paone, S., Baxter, A. A., Hulett, M. D., & Poon, I. K. H. (2019). Endothelial cell apoptosis and the role of endothelial cell-derived extracellular vesicles in the progression of atherosclerosis. Cellular and Molecular Life Sciences, 76, 1093–1106.
Vanhaverbeke, M., Gal, D., & Holvoet, P. (2017). Functional role of cardiovascular exosomes in myocardial injury and atherosclerosis. Advances in Experimental Medicine and Biology, 998, 45–58.
Huber, H. J., & Holvoet, P. (2015). Exosomes: Emerging roles in communication between blood cells and vascular tissues during atherosclerosis. Current Opinion in Lipidology, 26, 412–419.
Zhu, C., Li, L., Wang, Z., Irfan, M., & Qu, F. (2020). Recent advances of aptasensors for exosomes detection. Biosensors & Bioelectronics, 160, 112213.
Zhang, L., Gu, C., Wen, J., Liu, G., Liu, H., & Li, L. (2021). Recent advances in nanomaterial-based biosensors for the detection of exosomes. Analytical and Bioanalytical Chemistry, 413, 83–102.
Khodashenas, S., Khalili, S., & Forouzandeh Moghadam, M. (2019). A cell ELISA based method for exosome detection in diagnostic and therapeutic applications. Biotechnology Letters, 41, 523–531.
Pan, Y., Wang, L., Deng, Y., Wang, M., Peng, Y., Yang, J., & Li, G. (2020). A simple and sensitive method for exosome detection based on steric hindrance-controlled signal amplification. Chemical Communications (Cambridge, England), 56, 13768–13771.
Yang, F., Liao, X., Tian, Y. & Li, G. (2017). Exosome separation using microfluidic systems: size-based, immunoaffinity-based and dynamic methodologies. Biotechnol Journal, 12, 1600699
Ibsen, S. D., Wright, J., Lewis, J. M., Kim, S., Ko, S. Y., Ong, J., Manouchehri, S., Vyas, A., Akers, J., Chen, C. C., Carter, B. S., Esener, S. C., & Heller, M. J. (2017). Rapid isolation and detection of exosomes and associated biomarkers from plasma. ACS Nano, 11, 6641–6651.
Koritzinsky, E. H., Street, J. M., Star, R. A., & Yuen, P. S. (2017). Quantification of Exosomes. Journal of Cellular Physiology, 232, 1587–1590.
Zhao, X., Luo, C., Mei, Q., Zhang, H., Zhang, W., Su, D., Fu, W., & Luo, Y. (2020). Aptamer-cholesterol-mediated proximity ligation assay for accurate identification of exosomes. Analytical Chemistry, 92, 5411–5418.
Zhao, X., Yuan, Y., Liu, X., Mao, F., Xu, G., & Liu, Q. (2022). A versatile platform for sensitive and label-free identification of biomarkers through an Exo-III-assisted cascade signal amplification strategy. Analytical Chemistry, 94, 2298–2304.
Zhao, X., Zeng, L., Mei, Q., & Luo, Y. (2020). Allosteric probe-initiated wash-free method for sensitive extracellular vesicle detection through dual cycle-assisted CRISPR-Cas12a. ACS Sens, 5, 2239–2246.
Zhao, X., Zhang, W., Qiu, X., Mei, Q., Luo, Y., & Fu, W. (2020). Rapid and sensitive exosome detection with CRISPR/Cas12a. Analytical and Bioanalytical Chemistry, 412, 601–609.
Gong, S., Li, J., Pan, W., Li, N., & Tang, B. (2021). Duplex-specific nuclease-assisted CRISPR-Cas12a strategy for microRNA detection using a personal glucose meter. Analytical Chemistry, 93, 10719–10726.
Ahn, J. K., Kim, H. Y., Park, K. S., & Park, H. G. (2018). A personal glucose meter for label-free and washing-free biomolecular detection. Analytical Chemistry, 90, 11340–11343.
Chen, G. Y., Zhang, H., & Yang, F. Q. (2021). A simple and portable method for beta-glucosidase activity assay and its inhibitor screening based on a personal glucose meter. Analytica Chimica Acta, 1142, 19–27.
Acknowledgements
The authors thank the financial and technical support from The Third Affiliated Hospital of Chongqing Medical University.
Author information
Authors and Affiliations
Contributions
L.L financed the research. H.W. and L.L designed and wrote the manuscript. H.W. performed experiments. S.H, Z.X., D.X. and L.Y. assisted data analysis.
Corresponding author
Ethics declarations
Ethics Approval
Permission from the Institutional Animal Ethical Committee was received before making these experiments.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hu, W., Su, H., Zeng, X. et al. Exo-III Enzyme and DNAzyme-Assisted Dual Signal Recycles for Sensitive Analysis of Exosomes by Using Personal Glucose Meter. Appl Biochem Biotechnol 195, 861–870 (2023). https://doi.org/10.1007/s12010-022-04171-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-022-04171-5