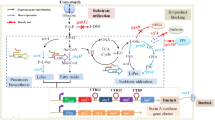
Abstract
The raw material cost of lactic acid fermentation accounts for the main part of the production cost, and this necessitates the exploration of the efficient use of cheap raw materials in lactic acid production. We compared the outcomes of the homologous expressions of xylose transporters (xylFGH, xylE, araE, and xylT), 6-phosphofructokinase (pfkA), fructose-bisphosphate aldolase (fbaA), and their co-expression in Lactococcus lactis IO-1 on lactic acid production using xylose as the raw material. We found that the production rate of lactic acid on xylose fermentation by L. lactis IO-1 overexpressing fbaA was the highest (14.42%). Among the xylose transporters investigated, XylT had the strongest xylose transport capacity in L. lactis IO-1, with an increase in the lactic acid production rate by 10.38%. The genes near the overexpression of fbaA or xylT in the metabolic pathway were more upregulated than the distant genes. The co-expression of fbaA and xylT increased the production rate of lactic acid by 27.84% on xylose fermentation by L. lactis IO-1. This work presents a novel strategy for the simultaneous enhancement of the expression of important genes at the beginning and midway of the xylose metabolic pathway of L. lactis IO-1, which could greatly improve the target production.
Graphical abstract





Similar content being viewed by others
Data Availability
All data generated or analyzed during this study are included in this published article.
References
Chen, H., Liu, J., Chang, X., Chen, D., Xue, Y., Liu, P., Lin, H., & Han, S. (2017). A review on the pretreatment of lignocellulose for high-value chemicals. Fuel Processing Technology, 160, 196–206.
Gu, Y., Li, J., Zhang, L., Chen, J., Niu, L., Yang, Y., Yang, S., & Jiang, W. (2009). Improvement of xylose utilization in Clostridium acetobutylicum via expression of the talA gene encoding transaldolase from Escherichia coli. Journal of Biotechnology, 143, 284–287.
Yanase, H., Sato, D., Yamamoto, K., Matsuda, S., Yamamoto, S., & Okamoto, K. (2007). Genetic engineering of Zymobacter palmae for production of ethanol from xylose. Applied and Environmental Microbiology, 73, 2592–2599.
Tanaka, K., Komiyama, A., Sonomoto, K., Ishizaki, A., Hall, S., & Stanbury, P. (2002). Two different pathways for D-xylose metabolism and the effect of xylose concentration on the yield coefficient of L-lactate in mixed-acid fermentation by the lactic acid bacterium Lactococcus lactis IO-1. Applied Microbiology Biotechnology, 60, 160–167.
Poorinmohammad, N., Hamedi, J., & Masoudi-Nejad, A. (2020). Genome-scale exploration of transcriptional regulation in the nisin Z producer Lactococcus lactis subsp. lactis IO-1. Scientific reports, 10(1), 1–13.
Tanaka, K., Katamune, K., & Ishizaki, A. (1993). Fermentative production of poly-β-hydroxybutyric acid from xylose by a two-stage culture method employing Lactococcus lactis IO-1 and Alcaligenes eutrophus. Biotechnology letters, 15(12), 1217–1222.
Kato, H., Shiwa, Y., Oshima, K., Machii, M., Araya-Kojima, T., Zendo, T., Shimizu-Kadota, M., Hattori, M., Sonomoto, K., & Yoshikawa, H. (2012). Complete genome sequence of Lactococcus lactis IO-1, a lactic acid bacterium that utilizes xylose and produces high levels of L-lactic acid. Journal of Bacteriology, 194(8), 2102–2103.
Nakahigashi, K., Toya, Y., Ishii, N., Soga, T., Hasegawa, M., Watanabe, H., Takai, Y., Honma, M., Mori, H., & Tomita, M. (2009). Systematic phenome analysis of Escherichia coli multiple-knockout mutants reveals hidden reactions in central carbon metabolism. Molecular Systems Biology, 5, 306.
Sirisansaneeyakul, S., Luangpipat, T., Vanichsriratana, W., Srinophakun, T., Chen, H. H. H., & Chisti, Y. (2007). Optimization of lactic acid production by immobilized Lactococcus lactis IO-1. Journal of Industrial Microbiology and Biotechnology, 34(5), 381.
Vonktaveesuk, P., Tonokawa, M., & Ishizaki, A. (1994). Stimulation of the rate of L-lactate fermentation using Lactococcus lactis IO-1 by periodic electrodialysis. Journal of fermentation and bioengineering, 77(5), 508–512.
Ishizaki, A., & Vonktaveesuk, P. (1996). Optimization of substrate feed for continuous production of lactic acid by Lactococcus lactis IO-1. Biotechnology letters, 18(10), 1113–1118.
Chaillou, S., Bor, Y.-C., Batt, C. A., Postma, P. W., & Pouwels, P. H. (1998). Molecular cloning and functional expression in Lactobacillus plantarum 80 of xylT, encoding the d-xylose–H+ symporter of Lactobacillus brevis. Applied and Environmental Microbiology, 64, 4720–4728.
Chaillou, S. P., Lokman, B. C., Leer, R. J., Posthuma, C., Postma, P. W., & Pouwels, P. H. (1998). Cloning, sequence analysis, and characterization of the genes involved in isoprimeverose metabolism in Lactobacillus pentosus. Journal of Bacteriology, 180, 2312–2320.
Qiu, Z., Gao, Q., & Bao, J. (2017). Constructing xylose-assimilating pathways in Pediococcus acidilactici for high titer D-lactic acid fermentation from corn stover feedstock. Bioresource Technology, 245, 1369–1376.
He, J., Liu, X., Xia, J., Xu, J., Xiong, P., & Qiu, Z. (2020). One-step utilization of non-detoxified pretreated lignocellulose for enhanced cellulolytic enzyme production using recombinant Trichoderma reesei RUT C30 carrying alcohol dehydrogenase and nicotinate phosphoribosyltransferase. Bioresource Technology, 310, 123458.
He, J., Sakaguchi, K., & Suzuki, T. (2012). Acquired tolerance to oxidative stress in Bifidobacterium longum 105-A via expression of a catalase gene. Applied Environmental Microbiology, 78, 2988–2990.
Diaz, C. A., Bennett, R. K., Papoutsakis, E. T., & Antoniewicz, M. R. (2019). Deletion of four genes in Escherichia coli enables preferential consumption of xylose and secretion of glucose. Metabolic Engineering, 52, 168–177.
Yuan, X., Wang, J., Lin, J., Yang, L., & Wu, M. (2019). Efficient production of xylitol by the integration of multiple copies of xylose reductase gene and the deletion of Embden–Meyerhof–Parnas pathway-associated genes to enhance NADPH regeneration in Escherichia coli. Journal of Industrial Microbiology and Biotechnology, 46, 1061–1069.
Westbrook, A. W., Ren, X., Oh, J., Moo-Young, M., & Chou, C. P. (2018). Metabolic engineering to enhance heterologous production of hyaluronic acid in Bacillus subtilis. Metabolic Engineering., 47, 401–413.
Yamamoto, S., Gunji, W., Suzuki, H., Toda, H., Suda, M., Jojima, T., Inui, M., & Yukawa, H. (2012). Overexpression of genes encoding glycolytic enzymes in Corynebacterium glutamicum enhances glucose metabolism and alanine production under oxygen deprivation conditions. Applied and environmental microbiology, 78(12), 4447–4457.
Guo, W., He, R., Ma, L., Jia, W., Li, D., & Chen, S. (2014). Construction of a constitutively expressed homo-fermentative pathway in Lactobacillus brevis. Applied Microbiology Biotechnology, 98, 6641–6650.
Tao, H., Gonzalez, R., Martinez, A., Rodriguez, M., Ingram, L., Preston, J., & Shanmugam, K. (2001). Engineering a homo-ethanol pathway in Escherichia coli: Increased glycolytic flux and levels of expression of glycolytic genes during xylose fermentation. Journal of Bacteriology, 183, 2979–2988.
Tarr, P. T., Tarling, E. J., Bojanic, D. D., Edwards, P. A., & Baldán, Á. (2009). Emerging new paradigms for ABCG transporters. Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids, 1791, 584–593.
Nataf, Y., Yaron, S., Stahl, F., Lamed, R., Bayer, E. A., Scheper, T.-H., Sonenshein, A. L., & Shoham, Y. (2009). Cellodextrin and laminaribiose ABC transporters in Clostridium thermocellum. Journal of Bacteriology., 191, 203–209.
Dunn, K. L., & Rao, C. V. (2014). Expression of a xylose-specific transporter improves ethanol production by metabolically engineered Zymomonas mobilis. Applied Microbiology Biotechnology, 98, 6897–6905.
Bueno, J. G. R., Borelli, G., Corrêa, T. L. R., Fiamenghi, M. B., José, J., de Carvalho, M., de Oliveira, L. C., Pereira, G. A., & Dos Santos, L. V. (2020). Novel xylose transporter Cs4130 expands the sugar uptake repertoire in recombinant Saccharomyces cerevisiae strains at high xylose concentrations. Biotechnology for Biofuels, 13, 1–20.
Zhao, Z., Xian, M., Liu, M., & Zhao, G. (2020). Biochemical routes for uptake and conversion of xylose by microorganisms. Biotechnology for Biofuels, 13, 1–12.
Jagtap, S. S., & Rao, C. V. (2018). Microbial conversion of xylose into useful bioproducts. Applied Microbiology Biotechnology, 102, 9015–9036.
Young, E., Poucher, A., Comer, A., Bailey, A., & Alper, H. (2011). Functional survey for heterologous sugar transport proteins, using Saccharomyces cerevisiae as a host. Applied and Environmental Microbiology, 77, 3311–3319.
Hasona, A., Kim, Y., Healy, F. G., Ingram, L., & Shanmugam, K. (2004). Pyruvate formate lyase and acetate kinase are essential for anaerobic growth of Escherichia coli on xylose. Journal of Bacteriology, 186, 7593–7600.
Griffith, J. K., Baker, M. E., Rouch, D. A., Page, M. G., Skurray, R. A., Paulsen, I. T., Chater, K. F., Baldwin, S. A., & Henderson, P. J. (1992). Membrane transport proteins: Implications of sequence comparisons. Current Opinion in Cell Biology, 4, 684–695.
Sedlak, M., & Ho, N. W. Y. (2004). Production of ethanol from cellulosic biomass hydrolysates using genetically engineered Saccharomyces yeast capable of cofermenting glucose and xylose. In M. Finkelstein, J. D. McMillan, B. H. Davison, & B. Evans (Eds.), Proceedings of the twenty-fifth symposium on biotechnology for fuels and chemicals held May 4–7, 2003, in Breckenridge, CO (pp. 403–416). Biotechnology for Fuels and Chemicals. Humana Press.
Chaillou, S., Pouwels, P. H., & Postma, P. W. (1999). Transport of D-xylose in Lactobacillus pentosus, Lactobacillus casei, and Lactobacillus plantarum: Evidence for a mechanism of facilitated diffusion via the phosphoenolpyruvate: Mannose phosphotransferase system. Journal of Bacteriology, 181, 4768–4773.
Desai, T. A., & Rao, C. V. (2010). Regulation of arabinose and xylose metabolism in Escherichia coli. Applied and Environmental Microbiology, 76, 1524–1532.
Funding
This work was supported by the National Natural Science Foundation of China (grant number 31200023; 22008083), and the Natural Science Foundation of Huaian City (HAB202054).
Author information
Authors and Affiliations
Contributions
Material preparation, data collection, and analysis were performed by Hanwen Zhang, Yuxiang Yang, and Wenyi Hou. Yejuan Qiu, Xiangqian Li, and Jianlong He designed the experiments and wrote the manuscript. Zhongyang Qiu, Jun Xia, and Xiaoyan Liu commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Qiu, Y., Qiu, Z., Xia, J. et al. Co-expression of Xylose Transporter and Fructose-Bisphosphate Aldolase Enhances the Utilization of Xylose by Lactococcus lactis IO-1. Appl Biochem Biotechnol 195, 816–831 (2023). https://doi.org/10.1007/s12010-022-04168-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-022-04168-0