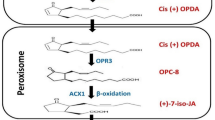
Abstract
Saline stress is the most limiting condition impacting the plant growth, development, and productivity. In this present study, jasmonic acid (JA) was used as a foliar spray on the rice seedlings grown under saline stress. Increase in photosynthetic pigments, anthocyanin, and total protein content was observed with JA treatment while NaCl showed reduction in biochemical constituents and enhanced antioxidant enzyme activity. The leaf cells of NaCl-treated seedlings accumulated more ROS and had more fragmented nuclei, whereas JA decreased the accumulation and fragmentation during saline stress. In NaCl treatment, gene expression analysis showed many fold upregulation in comparison with other treatments. The results suggest that JA acts as a promoter for growth, physiological, biochemical, and cellular contents, as well as ameliorate the effects of saline stress. The expression of genes demonstrated that saline stress may promote autophagy, which leads to autophagic cell death, and improve tolerance to saline stress in rice seedlings via the jasmonic acid signaling pathway. However, the mechanism by which jasmonate signaling induces autophagy and cell death is unknown and requires further exploration.






Similar content being viewed by others
Data Availability
Not applicable.
Code Availability
Not applicable.
References
Wasternack, C., & Hause, B. (2013). Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Annals of Botany, 111(6), 1021–1058
Santino, A., Taurino, M., De Domenico, S., Bonsegna, S., Poltronieri, P., Pastor, V., & Flors, V. (2013). Jasmonate signaling in plant development and defense response to multiple (a)biotic stresses. Plant Cell Reports. https://doi.org/10.1007/s00299-013-1441-2
El-Esawi, M. A. (2016). Jasmonic acid: Genetic pathway, signal transduction and action in plant development and defence. Biochemistry & Physiology: Open Access, 5(2), 1–2. https://doi.org/10.4172/2165-7890.1000E149
Qiu, Z., Guo, J., Zhu, A., Zhang, L., & Zhang, M. (2014). Exogenous jasmonic acid can enhance tolerance of wheat seedlings to salt stress. Ecotoxicology and Environmental Safety. https://doi.org/10.1016/j.ecoenv.2014.03.014
Munns, R. (2002). Comparative physiology of salt and water stress. Plant, Cell and Environment, 25(2), 239–250. https://doi.org/10.1046/J.0016-8025.2001.00808.X
Qin, H., & Huang, R. (2020). The phytohormonal regulation of Na+/K+ and reactive oxygen species homeostasis in rice salt response. Molecular Breeding, 40(5), 1–13. https://doi.org/10.1007/S11032-020-1100-6
Ma, X., Feng, F., Wei, H., Mei, H., Xu, K., Chen, S., & Luo, L. (2016). Genome-wide association study for plant height and grain yield in rice under contrasting moisture regimes. Frontiers in Plant Science, 7(NOVEMBER2016), 1801. https://doi.org/10.3389/FPLS.2016.01801/BIBTEX
Muranaka, S., Shimizu, K., & Kato, M. (2002). Ionic and osmotic effects of salinity on single-leaf photosynthesis in two wheat cultivars with different drought tolerance. Photosynthetica, 40(2), 201–207. https://doi.org/10.1023/A:1021337522431
Murphy, L. R., Kinsey, S. T., & Durako, M. J. (2003). Physiological effects of short-term salinity changes on Ruppia maritima. Aquatic Botany, 4(75), 293–309. https://doi.org/10.1016/S0304-3770(02)00206-1
Zhu, J. K. (2016). Abiotic stress signaling and responses in plants. Cell, 167(2), 313–324. https://doi.org/10.1016/J.CELL.2016.08.029
Miller, G., Suzuki, N., Ciftci-Yilmaz, S., & Mittler, R. (2010). Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant, Cell and Environment. https://doi.org/10.1111/j.1365-3040.2009.02041.x
He, C., & Klionsky, D. J. (2009). Regulation mechanisms and signaling pathways of autophagy. Annual review of genetics, 43, 67. https://doi.org/10.1146/Annurev-Genet-102808-114910
Mizushima, N., Yoshimori, T., & Ohsumi, Y. (2011). The role of Atg proteins in autophagosome formation. Annual review of cell and developmental biology, 27, 107–132. https://doi.org/10.1146/ANNUREV-CELLBIO-092910-154005
Gong, Q., Li, P., Ma, S., Indu Rupassara, S., & Bohnert, H. J. (2005). Salinity stress adaptation competence in the extremophile Thellungiella halophila in comparison with its relative Arabidopsis thaliana. The Plant journal : For cell and molecular biology, 44(5), 826–839. https://doi.org/10.1111/J.1365-313X.2005.02587.X
Liu, Y., Xiong, Y., & Bassham, D. C. (2009). Autophagy is required for tolerance of drought and salt stress in plants. Autophagy. https://doi.org/10.4161/auto.5.7.9290
Xia, K., Liu, T., Ouyang, J., Wang, R., Fan, T., & Zhang, M. (2011). Genome-wide identification, classification, and expression analysis of autophagy-associated gene homologues in rice (Oryza sativa L.). DNA research : an international journal for rapid publication of reports on genes and genomes, 18(5), 363–377. https://doi.org/10.1093/DNARES/DSR024
Zhai, Y., Guo, M., Wang, H., Lu, J., Liu, J., Zhang, C., & Lu, M. (2016). Autophagy, a conserved mechanism for protein degradation, responds to heat, and other abiotic stresses in Capsicum annuum L. Frontiers in Plant Science. https://doi.org/10.3389/fpls.2016.00131
Liao, C. Y., & Bassham, D. C. (2020). Combating stress: The interplay between hormone signaling and autophagy in plants. Journal of experimental botany, 71(5), 1723–1733. https://doi.org/10.1093/JXB/ERZ515
Gou, W., Li, X., Guo, S., Liu, Y., Li, F., & Xie, Q. (2019). Autophagy in plant: A new orchestrator in the regulation of the phytohormones homeostasis. International Journal of Molecular Sciences, 20(12), 2900.
Yoshimoto, K., Jikumaru, Y., Kamiya, Y., Kusano, M., Consonni, C., Panstruga, R., & Shirasu, K. (2009). Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. The Plant Cell, 21(9), 2914–2927. https://doi.org/10.1105/TPC.109.068635
Mackinney, G. (1941). Absorption of light by chlorophyll. Absorption of Light by Chlorophyll, 140, 315–323.
Laxmi, A., Paul, L. K., Raychaudhuri, A., Peters, J. L., & Khurana, J. P. (2006). Arabidopsis cytokinin-resistant mutant, cnr1, displays altered auxin responses and sugar sensitivity. Plant Molecular Biology, 62(3), 409–425. https://doi.org/10.1007/s11103-006-9032-z
Lowry, O. H., Rosebrough, N. J., Farr, A. L., & Randall, R. J. (1951). Protein measurement with the Folin phenol reagent. The Journal of Biological Chemistry, 193(1), 265–75.
Idrees, M., Naeem, M., Aftab, T., Khan, M. M. A.,& Moinuddin. (2011). Salicylic acid mitigates salinity stress by improving antioxidant defence system and enhances vincristine and vinblastine alkaloids production in periwinkle [Catharanthus roseus (L.) G. Don]. Acta Physiologiae Plantarum, 33(3), 987–999.https://doi.org/10.1007/s11738-010-0631-6
Chandlee, J. M., & Scandalios, J. G. (1984). Analysis of variants affecting the catalase developmental program in maize scutellum. Theoretical and Applied Genetics. https://doi.org/10.1007/BF00262543
Beauchamp, C., & Fridovich, I. (1971). Superoxide dismutase: Improved assays and an assay applicable to acrylamide gels. Analytical biochemistry, 44(1), 276–87. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/4943714. Accessed 01.12.2018
Khan, M. S., Pandey, M. K., & Hemalatha, S. (2018). Comparative studies on the role of organic biostimulant in resistant and susceptible cultivars of rice grown under saline stress - organic biostimulant alleviate saline stress in tolerant and susceptible cultivars of rice. Journal of Crop Science and Biotechnology, 21(5), 459–467.
Sudhir, P., & Murthy, S. D. S. (2004). Effects of salt stress on basic processes of photosynthesis. Photosynthetica. https://doi.org/10.1007/S11099-005-0001-6
Qados, A. A. M. S. (2011). Effect of salt stress on plant growth and metabolism of bean plant Vicia faba (L.). Journal of the Saudi Society of Agricultural Sciences, 10(1), 7–15.
Ayala-Astorga, G. I., & Alcaraz-Meléndez, L. (2010). Salinity effects on protein content, lipid peroxidation, pigments, and proline in Paulownia imperialis (Siebold & Zuccarini) and Paulownia fortunei (Seemann & Hemsley) grown in vitro. Electronic Journal of Biotechnology. https://doi.org/10.2225/vol13-issue5-fulltext-13
Qiu, Z. B., Guo, J. L., Zhu, A. J., Zhang, L., & Zhang, M. M. (2014). Exogenous jasmonic acid can enhance tolerance of wheat seedlings to salt stress. Ecotoxicology and environmental safety, 104(1), 202–208. https://doi.org/10.1016/J.ECOENV.2014.03.014
Parida, A. K., Das, A. B., & Mohanty, P. (2004). Defense potentials to NaCl in a mangrove, Bruguiera parviflora: Differential changes of isoforms of some antioxidative enzymes. Journal of Plant Physiology, 161(5), 531–542.
Jithesh, M. N., Prashanth, S. R., Sivaprakash, K. R., & Parida, A. K. (2006). Antioxidative response mechanisms in halophytes: Their role in stress defence. Journal of Genetics, 85(3), 237–254. https://doi.org/10.1007/BF02935340
Matamoros, M. A. (2003). Biochemistry and molecular biology of antioxidants in the rhizobia-legume symbiosis. PLANT PHYSIOLOGY. https://doi.org/10.1104/pp.103.025619
Anjum, S. A., Xie, X., & Wang, L. (2011). Morphological, physiological and biochemical responses of plants to drought stress. African Journal of Agricultural Research. https://doi.org/10.5897/AJAR10.027
Li, Q., Yang, A., & Zhang, W. H. (2017). Comparative studies on tolerance of rice genotypes differing in their tolerance to moderate salt stress. BMC Plant Biology. https://doi.org/10.1186/s12870-017-1089-0
Yazdani, M., & Mahdieh, M. (2012). Salinity induced apoptosis in root meristematic cells of rice, 2(1), 2–5. https://doi.org/10.7763/IJBBB.2012.V2.66
Browse, J. (2009). Jasmonate passes muster: A receptor and targets for the defense hormone. Annual Review of Plant Biology. https://doi.org/10.1146/annurev.arplant.043008.092007
Santino, A., Taurino, M., De Domenico, S., Bonsegna, S., Poltronieri, P., Pastor, V., & Flors, V. (2013). Jasmonate signaling in plant development and defense response to multiple (a)biotic stresses. Plant cell reports, 32(7), 1085–1098. https://doi.org/10.1007/S00299-013-1441-2
Svyatyna, K., & Riemann, M. (2012). Light-dependent regulation of the jasmonate pathway. Protoplasma. https://doi.org/10.1007/s00709-012-0409-3
Lu, Z., Liu, D., & Liu, S. (2007). Two rice cytosolic ascorbate peroxidases differentially improve salt tolerance in transgenic Arabidopsis. Plant Cell Reports. https://doi.org/10.1007/s00299-007-0395-7
Pérez-Clemente, R. M., Vives, V., Zandalinas, S. I., López-Climent, M. F., Muñoz, V., & Gómez-Cadenas, A. (2013). Biotechnological approaches to study plant responses to stress. BioMed Research International. https://doi.org/10.1155/2013/654120
Im, J. H., Lee, H., Kim, J., Kim, H. B., Seyoung, K., Kim, B. M., & An, C. S. (2012). A salt stress-activated mitogen-activated protein kinase in soybean is regulated by phosphatidic acid in early stages of the stress response. Journal of Plant Biology. https://doi.org/10.1007/s12374-011-0036-8
Yousfi, F. E., Makhloufi, E., Marande, W., Ghorbel, A. W., Bouzayen, M., & Bergès, H. (2017). Comparative analysis of WRKY genes potentially involved in salt stress responses in Triticum turgidum L. ssp. durum. Frontiers in Plant Science, 7, 2034. https://doi.org/10.3389/fpls.2016.02034
Lai, Z., Wang, F., Zheng, Z., Fan, B., & Chen, Z. (2011). A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant Journal. https://doi.org/10.1111/j.1365-313X.2011.04553.x
Akimoto-Tomiyama, C., Sakata, K., Yazaki, J., Nakamura, K., Fujii, F., Shimbo, K., & Minami, E. (2003). Rice gene expression in response to N-acetylchitooligosaccharide elicitor: Comprehensive analysis by DNA microarray with randomly selected ESTs. Plant Molecular Biology. https://doi.org/10.1016/j.athoracsur.2017.01.116
Ramamoorthy, R., Jiang, S. Y., Kumar, N., Venkatesh, P. N., & Ramachandran, S. (2008). A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments. Plant and Cell Physiology. https://doi.org/10.1093/pcp/pcn061
Miao, Y., Laun, T. M., Smykowski, A., & Zentgraf, U. (2007). Arabidopsis MEKK1 can take a short cut: It can directly interact with senescence-related WRKY53 transcription factor on the protein level and can bind to its promoter. Plant Molecular Biology. https://doi.org/10.1007/s11103-007-9198-z
Besseau, S., Li, J., & Palva, E. T. (2012). WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. Journal of Experimental Botany. https://doi.org/10.1093/jxb/err450
Tsugane, K., Kobayashi, K., Niwa, Y., Ohba, Y., Wada, K., & Kobayashi, H. (1999). A recessive Arabidopsis mutant that grows photoautotrophically under salt stress shows enhanced active oxygen detoxification. The Plant Cell, 11(7), 1195–1206. https://doi.org/10.1105/TPC.11.7.1195
Slavikova, S., Ufaz, S., Avin-Wittenberg, T., Levanony, H., & Galili, G. (2008). An autophagy-associated Atg8 protein is involved in the responses of Arabidopsis seedlings to hormonal controls and abiotic stresses. Journal of Experimental Botany. https://doi.org/10.1093/jxb/ern244
Bassham, D. C., Laporte, M., Marty, F., Moriyasu, Y., Ohsumi, Y., Olsen, L. J., & Yoshimoto, K. (2006). Autophagy in development and stress responses of plants. Autophagy. https://doi.org/10.4161/auto.2092
Shibuya, K., Niki, T., & Ichimura, K. (2013). Pollination induces autophagy in petunia petals via ethylene. Journal of Experimental Botany, 64(4), 1111. https://doi.org/10.1093/JXB/ERS395
Funding
This work was supported by B. S. Abdur Rahman Crescent Institute of Science and Technology, Chennai, TN, India.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Khan, M.S., Hemalatha, S. Autophagy and Programmed Cell Death Are Critical Pathways in Jasmonic Acid Mediated Saline Stress Tolerance in Oryza sativa. Appl Biochem Biotechnol 194, 5353–5366 (2022). https://doi.org/10.1007/s12010-022-04032-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-022-04032-1