Abstract
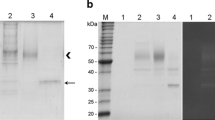
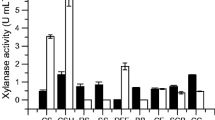
Xylanase plays a vital role in the efficient utilization of xylan, which accounts for up to 30% of plant dry matter. However, the production cost of xylanase remains high, and the enzymatic characteristics of xylanases of most microorganisms are not suitable for industrial production. Therefore, it is of great significance to discover and develop new and efficient xylanases. In this study, the xylanase gene TAX1 (672 bp cDNA) was cloned from Trichoderma atroviride 3.3013 and expressed in Pichia pastoris. The TAX1 gene encoded a 223-amino acid protein (TAX1) with a molecular weight of 24.2 kDa which showed high similarity to glycoside hydrolase family 11. Enzyme activity assay verified that the recombinant xylanase TAX1 had optimal activity (215.3 IU/mL) at 50°C and pH 6.0. Stable working conditions were measured as pH 4.0–7.0 and 40–60°C. By adding Zn2+, the relative enzymatic activity of recombinant TAX1 was enhanced by 26%. The recombinant xylanase showed high activity toward birchwood xylan and corn stover. The Km and Kcat for xylan and corn stover were 0.36 mg/mL and 0.204 S−1 and 0.48 mg/mL and 0.149 S−1, respectively. The enzymatic activity of the TAX1 produced by P. pastoris was about 2.4–4 times higher that directly isolated from T. atroviride, so engineered P. pastoris for xylanase production could be an ideal candidate for industrial enzyme production.






Similar content being viewed by others
Availability of data and materials
The data is included in the article and supporting materials. Additional information can be acquired by contacting the corresponding author.
References
Bailey, M. J., Biely, P., & Poutanen, K. (1992). Interlaboratory testing of methods for assay of xylanase activity. Journal of Biotechnology, 23(3), 257–270.
Bhardwaj, N., Kumar, B., & Verma, P. (2019). A detailed overview of xylanases: An emerging biomolecule for current and future prospective. Bioresources and Bioprocessing, 6(1), 40.
Biely, P. (1985). Microbial xylanolytic systems. Trends in Biotechnology, 3(11), 286–290.
Biely, P., Kremnický, L., Alföldi, J., & Tenkanen, M. (1994). Stereochemistry of the hydrolysis of glycosidic linkage by endo-β-1,4-xylanases of Trichoderma reesei. FEBS Letters, 356(1), 137–140.
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry, 72(1-2), 248–254.
Buckholz, R. G., & Gleeson, M. A. G. (1991). Yeast systems for the commercial production of heterologous proteins. Bio/Technology, 9(11), 1067–1072.
Cornejo, A., Alegria-Dallo, I., García-Yoldi, Í., Sarobe, Í., Sánchez, D., Otazu, E., Funcia, I., Gil, M. J., & Martínez-Merino, V. (2019). Pretreatment and enzymatic hydrolysis for the efficient production of glucose and furfural from wheat straw, pine and poplar chips. Bioresource Technology, 288, 121583.
Covian, R., Chess, D., & Balaban, R. S. (2012). Continuous monitoring of enzymatic activity within native electrophoresis gels: Application to mitochondrial oxidative phosphorylation complexes. Analytical Biochemistry, 431(1), 30–39.
Duncan, D. B. (1955). Multiple range and multiple F tests. Biometrics, 11(1), 1–42.
Fang, W., Gao, H., Cao, Y., & Shan, A. (2014). Cloning and expression of a xylanase xynB from Aspergillus niger IA-001 in Pichia pastoris. Journal of Basic Microbiology, 54(S1), S190–S199.
Han, H., You, S., Zhu, B., Fu, X., Sun, B., Qiu, J., Yu, C., Chen, L., Peng, R., & Yao, Q. (2015). Characterization and high expression of recombinant Ustilago maydis xylanase in Pichia pastoris. Biotechnology Letters, 37(3), 697–703.
He, J., Yu, B., Zhang, K., Ding, X., & Chen, D. (2009). Expression of a Trichoderma reesei β-xylanase gene in Escherichia coli and activity of the enzyme on fiber-bound substrates. Protein Expression and Purification, 67, 1–6.
He, J., Yu, B., Zhang, K., Ding, X., & Chen, D. (2009). Expression of endo-1, 4-beta-xylanase from Trichoderma reesei in Pichia pastorisand functional characterization of the produced enzyme. BMC Biotechnology, 9(1), 56.
He, J., Tang, F., Chen, D., Yu, B., Luo, Y., Zheng, P., Mao, X., Yu, J., & Yu, F. (2019). Design, expression and functional characterization of a thermostable xylanase from Trichoderma reesei. PLoS One, 14(1), e0210548.
Jiang, Z. Q., Deng, W., Zhu, Y. P., Li, L. T., Sheng, Y. J., & Hayashi, K. (2004). The recombinant xylanase B of Thermotoga maritima is highly xylan specific and produces exclusively xylobiose from xylans, a unique character for industrial applications. Journal of Molecular Catalysis B: Enzymatic, 27(4-6), 207–213.
Julien, C. (2006). Production of humanlike recombinant proteins in Pichia pastoris from expression vector to fermentation strategy. Bioprocess International, 22–30.
Korkmaz, M. N., Ozdemir, S. C., & Uzel, A. (2017). Xylanase production from marine derived Trichoderma pleuroticola 08ÇK001 strain isolated from Mediterranean coastal sediments. Journal of Basic Microbiology, 57(10), 839–851.
Kulkarni, N., Shendye, A., & Rao, M. (1999). Molecular and biotechnological aspects of xylanases. FEMS Microbiology Reviews, 23(4), 411–456.
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227(5259), 680–685.
Li, Y.-y., Zhong, K.-x., Hu, A.-h., Liu, D.-n., Chen, L.-z., & Xu, S.-d. (2015). High-level expression and characterization of a thermostable xylanase mutant from Trichoderma reesei in Pichia pastoris. Protein Expression and Purification, 108, 90–96.
Li, Z., Zhang, X., Li, C., Kovalevsky, A., & Wan, Q. (2020). Studying the role of a single mutation of a family 11 glycoside hydrolase using high-resolution X-ray crystallography. The Protein Journal, 39(6), 671–680.
Livak, K. J., & Schmittgen, T. D. J. m. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods, 25(4), 402–408.
López, G., Bañares-Hidalgo, A., & Estrada, P. (2011). Xylanase II from Trichoderma reesei QM 9414: Conformational and catalytic stability to Chaotropes, Trifluoroethanol, and pH changes. Journal of Industrial Microbiology & Biotechnology, 38(1), 113–125.
Lu, F., Lu, M., Lu, Z., Bie, X., Zhao, H., & Wang, Y. (2008). Purification and characterization of xylanase from Aspergillus ficuum AF-98. Bioresource Technology, 99(13), 5938–5941.
Miller, G. L. J. A. c. (1959). Use of dinitrosalicylic acid reagent for determination of reducing sugar. Analytical Chemistry, 31(3), 426–428.
Moukouli, M., Topakas, E., & Christakopoulos, P. (2011). Cloning and optimized expression of a GH-11 xylanase from Fusarium oxysporum in Pichia pastoris. New Biotechnology, 28(4), 369–374.
Nair, S. G., Sindhu, R., & Shashidhar, S. (2008). Purification and biochemical characterization of two xylanases from Aspergillus sydowii SBS 45. Applied Biochemistry and Biotechnology, 149(3), 229–243.
Ostadjoo, S., Hammerer, F., Dietrich, K., Dumont, M.-J., Friščić, T., & Auclair, K. (2019). Efficient enzymatic hydrolysis of biomass hemicellulose in the absence of bulk water. Molecules, 24(23), 4206.
Romanos, M. A., Scorer, C. A., & Clare, J. J. J. Y. (1992). Foreign gene expression in yeast: A review. Yeast, 8(6), 423–488.
Ruanglek, V., Sriprang, R., Ratanaphan, N., Tirawongsaroj, P., Chantasigh, D., Tanapongpipat, S., Pootanakit, K., & Eurwilaichitr, L. (2007). Cloning, expression, characterization, and high cell-density production of recombinant endo-1,4-β-xylanase from Aspergillus niger in Pichia pastoris. Enzyme and Microbial Technology, 41(1-2), 19–25.
Sriyapai, T., Somyoonsap, P., Matsui, K., Kawai, F., & Chansiri, K. (2011). Cloning of a thermostable xylanase from Actinomadura sp. S14 and its expression in Escherichia coli and Pichia pastoris. Journal of Bioscience and Bioengineering, 111(5), 528–536.
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., & Higgins, D. G. (1997). The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 25(24), 4876–4882.
Waeonukul, R., Pason, P., Kyu, K. L., Sakka, K., Kosug, A., Mori, Y., & Ratanakhanokchai, K. J. J. o. m. and biotechnology. (2009). Cloning, sequencing, and expression of the gene encoding a multidomain endo-β-1,4-xylanase from Paenibacillus curdlanolyticus B-6, and characterization of the recombinant enzyme. Journal of Microbiology and Biotechnology, 19, 277–285.
Wei, Q. (1999). The guide of molecular biology experiment. Higher Education Publishing.
Yang, S. Q., Yan, Q. J., Jiang, Z. Q., Li, L. T., Tian, H. M., & Wang, Y. Z. (2006). High-level of xylanase production by the thermophilic Paecilomyces themophila J18 on wheat straw in solid-state fermentation. Bioresource Technology, 97(15), 1794–1800.
Yang, Y., Shan, K., Ping, L., & Lu, J. J. A. J. o. B. (2008). Cloning, sequencing and expression of a novel xylanase cDNA from a newly isolated Aspergillus awamori in Pichia pastoris. African Journal of Biotechnology, 7, 4251–4259.
Funding
This work was partially supported by funds provided by the Natural Science Foundation of Heilongjiang Province (C2015002 and LH2020C011) and the Postdoctoral Scientific Research Developmental Fund of Heilongjiang Province (LBH-Q18023).
Author information
Authors and Affiliations
Contributions
JC and XC performed research and wrote the paper. JF designed the study and wrote the paper. XH designed the study, provided funding, and edited the paper. RL analyzed the data and edited the paper. XS, QL, and DL performed research.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
No conflicts, informed consent, and human or animal rights applicable.
Consent for publication
All authors agree to publish this article in Applied Microbiology and Biotechnology.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 251 kb)
Rights and permissions
About this article
Cite this article
Cai, J., Chen, XL., Fan, JX. et al. Cloning and Heterologous Expression of a Novel Xylanase Gene TAX1 from Trichoderma atroviride and Its Application in the Deconstruction of Corn Stover. Appl Biochem Biotechnol 193, 3029–3044 (2021). https://doi.org/10.1007/s12010-021-03582-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-021-03582-0