Abstract
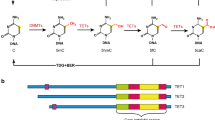
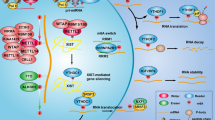
Cytosine methylation is a well-explored epigenetic modification mediated by DNA methyltransferases (DNMTs) which are considered “methylation writers”; cytosine methylation is a reversible process. The process of removal of methyl groups from DNA remained unelucidated until the discovery of ten-eleven translocation (TET) proteins which are now considered “methylation editors.” TET proteins are a family of Fe(II) and alpha-ketoglutarate-dependent 5-methyl cytosine dioxygenases—they convert 5-methyl cytosine to 5-hydroxymethyl cytosine, and to further oxidized derivatives. In humans, there are three TET paralogs with tissue-specific expression, namely TET1, TET2, and TET3. Among the TETs, TET2 is highly expressed in hematopoietic stem cells where it plays a pleiotropic role. The paralogs also differ in their structure and DNA binding. TET2 lacks the CXXC domain which mediates DNA binding in the other paralogs; thus, TET2 requires interactions with other proteins containing DNA-binding domains for effectively binding to DNA to bring about the catalysis. In addition to its role as methylation editor of DNA, TET2 also serves as methylation editor of RNA. Thus, TET2 is involved in epigenetics as well as epitranscriptomics. TET2 mutations have been found in various malignant hematological disorders like acute myeloid leukemia, and non-malignant hematological disorders like myelodysplastic syndromes. Increasing evidence shows that TET2 plays an important role in the non-hematopoietic system as well. Hepatocellular carcinoma, gastric cancer, prostate cancer, and melanoma are some non-hematological malignancies in which a role of TET2 has been implicated. Loss of TET2 is also associated with atherosclerotic vascular lesions and endometriosis. The current review elaborates on the role of structure, catalysis, physiological functions, pathological alterations, and methods to study TET2, with specific emphasis on epigenomics and epitranscriptomics.





Similar content being viewed by others
Data Availability
Data and materials will be freely available.
References
Prokhortchouk, E., & Defossez, P.-A. (2008). The cell biology of DNA methylation in mammals. Biochimica et Biophysica Acta, Molecular Cell Research, 1783(11), 2167–2173.
Jeltsch, A., Ehrenhofer-Murray, A., Jurkowski, T. P., Lyko, F., Reuter, G., Ankri, S., et al. (2017). Mechanism and biological role of Dnmt2 in nucleic acid methylation. RNA Biology, 14(9), 1108–1123.
Shimbo, T., & Wade, P. A. (2016). Proteins that read dna methylation. Advances in Experimental Medicine and Biology, 945, 303–320.
Du, Q., Luu, P.-L., Stirzaker, C., & Clark, S. J. (2015). Methyl-CpG-binding domain proteins: Readers of the epigenome. Epigenomics., 7(6), 1051–1073.
Martín Caballero, I., Hansen, J., Leaford, D., Pollard, S., & Hendrich, B. D. (2009). The methyl-CpG binding proteins Mecp2, Mbd2 and Kaiso are dispensable for mouse embryogenesis, but play a redundant function in neural differentiation. PLoS One, 4(1), e4315.
Vaughan, R. M., Dickson, B. M., Cornett, E. M., Harrison, J. S., Kuhlman, B., & Rothbart, S. B. (2018). Comparative biochemical analysis of UHRF proteins reveals molecular mechanisms that uncouple UHRF2 from DNA methylation maintenance. Nucleic Acids Research, 46(9), 4405–4416.
Clouaire, T., & Stancheva, I. (2008). Methyl-CpG binding proteins: Specialized transcriptional repressors or structural components of chromatin? Cellular and Molecular Life Sciences: CMLS, 65(10), 1509–1522.
Ooi, S. K. T., & Bestor, T. H. (2008). The colorful history of active DNA demethylation. Cell., 133(7), 1145–1148.
Ehrlich, M. (2009). DNA hypomethylation in cancer cells. Epigenomics., 1(2), 239–259.
Wu, S. C., & Zhang, Y. (2010). Active DNA demethylation: Many roads lead to Rome. Nature Reviews. Molecular Cell Biology, 11(9), 607–620.
Tahiliani, M., Koh, K. P., Shen, Y., Pastor, W. A., Bandukwala, H., Brudno, Y., et al. (2009). Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science., 324(5929), 930–935.
Lorsbach, R. B., Moore, J., Mathew, S., Raimondi, S. C., Mukatira, S. T., & Downing, J. R. (2003). TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23). Leukemia., 17(3), 637–641.
Pastor, W. A., Aravind, L., & Rao, A. (2013). TETonic shift: Biological roles of TET proteins in DNA demethylation and transcription. Nature Reviews. Molecular Cell Biology, 14(6), 341–356.
Hu, L., Li, Z., Cheng, J., Rao, Q., Gong, W., Liu, M., et al. (2013). Crystal structure of TET2-DNA complex: Insight into TET-mediated 5mC oxidation. Cell., 155(7), 1545–1555.
Bussaglia, E., Antón, R., Nomdedéu, J. F., & Fuentes-Prior, P. (2019). TET2 missense variants in human neoplasia. A proposal of structural and functional classification. Molecular Genetics & Genomic Medicine, 7(7), e00772.
IntEnz - EC 1.14.11.n2 [Internet]. Available from: https://www.ebi.ac.uk/intenz/query?q=1.14.11.n2&submit=Search. Accessed 22 Oct 2019.
Iyer, L. M., Tahiliani, M., Rao, A., & Aravind, L. (2009). Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle, 8(11), 1698–1710.
Liu, M. Y., Torabifard, H., Crawford, D. J., DeNizio, J. E., Cao, X.-J., Garcia, B. A., et al. (2017). Mutations along a TET2 active site scaffold stall oxidation at 5-hydroxymethylcytosine. Nature Chemical Biology, 13(2), 181–187.
Borst, P., & Sabatini, R. (2008). Base J: Discovery, biosynthesis, and possible functions. Annual Review of Microbiology, 62(1), 235–251.
Wu, H., D’Alessio, A. C., Ito, S., Xia, K., Wang, Z., Cui, K., et al. (2011). Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature., 473(7347), 389–393.
Ko, M., An, J., Bandukwala, H. S., Chavez, L., Äijö, T., Pastor, W. A., et al. (2013). Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature., 497(7447), 122–126.
Iyer, L. M., Abhiman, S., & Aravind, L. (2011). Chapter 2-Natural history of eukaryotic DNA methylation systems. In X. Cheng & R. M. Blumenthal (Eds.), Progress in molecular biology and translational science [Internet] (p. 25–104). Academic Press. (Modifications of Nuclear DNA and its Regulatory Proteins; vol. 101). Available from: http://www.sciencedirect.com/science/article/pii/B9780123876850000020. Accessed 27 Oct 2019.
Davey, N. E. (2019). The functional importance of structure in unstructured protein regions. Current Opinion in Structural Biology, 56, 155–163.
Ravichandran, M., Lei, R., Tang, Q., Zhao, Y., Lee, J., Ma, L., et al. (2019). Rinf regulates pluripotency network genes and Tet enzymes in embryonic stem cells. Cell Reports, 28(8), 1993–2003.e5.
Zhou, L., Ren, M., Zeng, T., Wang, W., Wang, X., Hu, M., et al. (2019). TET2-interacting long noncoding RNA promotes active DNA demethylation of the MMP-9 promoter in diabetic wound healing. Cell Death & Disease, 10(11), 813.
Delhommeau, F., Dupont, S., James, C., Masse, A., le Couedic, J. P., Valle, V. D., et al. (2008). TET2 is a novel tumor suppressor gene inactivated in myeloproliferative neoplasms: Identification of a pre-JAK2 V617F event. Blood, 112(11), lba-3-lba-3.
Jankowska, A. M., Szpurka, H., Tiu, R. V., Makishima, H., Afable, M., Huh, J., et al. (2009). Loss of heterozygosity 4q24 and TET2 mutations associated with myelodysplastic/myeloproliferative neoplasms. Blood., 113(25), 6403–6410.
Tefferi, A., Pardanani, A., Lim, K.-H., Abdel-Wahab, O., Lasho, T., Patel, J., et al. (2009). TET2 mutations and their clinical correlates in polycythemia vera, essential thrombocythemia and myelofibrosis. Leukemia., 23(5), 905–911.
Abdel-Wahab, O., Mullally, A., Hedvat, C., Garcia-Manero, G., Patel, J., Wadleigh, M., et al. (2009). Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood., 114(1), 144–147.
Tefferi, A., Levine, R. L., Lim, K.-H., Abdel-Wahab, O., Lasho, T. L., Patel, J., et al. (2009). Frequent TET2 mutations in systemic mastocytosis: Clinical, KITD816V and FIP1L1-PDGFRA correlates. Leukemia., 23(5), 900–904.
Mullighan, C. G. (2009). TET2 mutations in myelodysplasia and myeloid malignancies. Nature Genetics, 41(7), 766–767.
Verma, N., Pan, H., Doré, L. C., Shukla, A., Li, Q. V., Pelham-Webb, B., et al. (2018). TET proteins safeguard bivalent promoters from de novo methylation in human embryonic stem cells. Nature Genetics, 50(1), 83–95.
Koh, K. P., Yabuuchi, A., Rao, S., Huang, Y., Cunniff, K., Nardone, J., et al. (2011). Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell, 8(2), 200–213.
Ito, S., D’Alessio, A. C., Taranova, O. V., Hong, K., Sowers, L. C., & Zhang, Y. (2010). Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature., 466(7310), 1129–1133.
Uhlén, M., Fagerberg, L., Hallström, B. M., Lindskog, C., Oksvold, P., Mardinoglu, A., et al. (2015). Tissue-based map of the human proteome. Science, 347(6220), 1260419. Data available from https://www.proteinatlas.org/ENSG00000168769-TET2. Accessed 22 Oct 2019.
Ko, M., & Rao, A. (2011). TET2: Epigenetic safeguard for HSC. Blood., 118(17), 4501–4503.
Solary, E., Bernard, O. A., Tefferi, A., Fuks, F., & Vainchenker, W. (2014). The ten-eleven translocation-2 (TET2) gene in hematopoiesis and hematopoietic diseases. Leukemia., 28(3), 485–496.
Jaiswal, S., Fontanillas, P., Flannick, J., Manning, A., Grauman, P. V., Mar, B. G., et al. (2014). Age-related clonal hematopoiesis associated with adverse outcomes. The New England Journal of Medicine, 371(26), 2488–2498.
Fuster, J. J., MacLauchlan, S., Zuriaga, M. A., Polackal, M. N., Ostriker, A. C., Chakraborty, R., et al. (2017). Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science, 355(6327), 842–847.
Carrillo-Jimenez, A., Deniz, Ö., Niklison-Chirou, M. V., Ruiz, R., Bezerra-Salomão, K., Stratoulias, V., et al. (2019). TET2 regulates the neuroinflammatory response in microglia. Cell Reports, 29(3), 697–713.e8.
Cakouros, D., Hemming, S., Gronthos, K., Liu, R., Zannettino, A., Shi, S., et al. (2019). Specific functions of TET1 and TET2 in regulating mesenchymal cell lineage determination. Epigenetics & Chromatin, 12(1), 3.
Mi, Y., Gao, X., Dai, J., Ma, Y., Xu, L., & Jin, W. A. (2015). novel function of TET2 in CNS: Sustaining neuronal survival. International Journal of Molecular Sciences, 16(9), 21846–21857.
Yue, X., Lio, C.-W. J., Samaniego-Castruita, D., Li, X., & Rao, A. (2019). Loss of TET2 and TET3 in regulatory T cells unleashes effector function. Nature Communications, 10(1), 1–14.
Hahn, M. A., Qiu, R., Wu, X., Li, A. X., Zhang, H., Wang, J., et al. (2013). Dynamics of 5-hydroxymethylcytosine and chromatin marks in mammalian neurogenesis. Cell Reports, 3(2), 291–300.
Liu, R., Jin, Y., Tang, W. H., Qin, L., Zhang, X., Tellides, G., et al. (2013). Ten-eleven translocation-2 (TET2) is a master regulator of smooth muscle cell plasticity. Circulation., 128(18), 2047–2057.
Rasmussen, K. D., & Helin, K. (2016). Role of TET enzymes in DNA methylation, development, and cancer. Genes & Development, 30(7), 733–750.
Globisch, D., Münzel, M., Müller, M., Michalakis, S., Wagner, M., Koch, S., et al. (2010). Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS ONE, 5(12), e15367.
Mellén, M., Ayata, P., Dewell, S., Kriaucionis, S., & Heintz, N. (2012). MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell., 151(7), 1417–1430.
Iurlaro, M., Ficz, G., Oxley, D., Raiber, E.-A., Bachman, M., Booth, M. J., et al. (2013). A screen for hydroxymethylcytosine and formylcytosine binding proteins suggests functions in transcription and chromatin regulation. Genome Biology, 14(10), R119.
Chen, R., Zhang, Q., Duan, X., York, P., Chen, G.-D., Yin, P., et al. (2017). The 5-hydroxymethylcytosine (5hmC) reader UHRF2 is required for normal levels of 5hmC in mouse adult brain and spatial learning and memory. The Journal of Biological Chemistry, 292(11), 4533–4543.
Ito, S., Shen, L., Dai, Q., Wu, S. C., Collins, L. B., Swenberg, J. A., et al. (2011). Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science., 333(6047), 1300–1303.
Hu, L., Lu, J., Cheng, J., Rao, Q., Li, Z., Hou, H., et al. (2015). Structural insight into substrate preference for TET-mediated oxidation. Nature., 527(7576), 118–122.
Spruijt, C. G., Gnerlich, F., Smits, A. H., Pfaffeneder, T., Jansen, P. W. T. C., Bauer, C., et al. (2013). Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell., 152(5), 1146–1159.
Bonev, B., & Cavalli, G. (2016). Organization and function of the 3D genome. Nature Reviews. Genetics, 17(11), 661–678.
Ong, C.-T., & Corces, V. G. (2014). CTCF: An architectural protein bridging genome topology and function. Nature Reviews. Genetics, 15(4), 234–246.
Kim, T. H., Abdullaev, Z. K., Smith, A. D., Ching, K. A., Loukinov, D. I., Green, R. D., et al. (2007). Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell., 128(6), 1231–1245.
Marina, R. J., Sturgill, D., Bailly, M. A., Thenoz, M., Varma, G., Prigge, M. F., et al. (2016). TET-catalyzed oxidation of intragenic 5-methylcytosine regulates CTCF-dependent alternative splicing. The EMBO Journal, 35(3), 335–355.
Nanan, K. K., Sturgill, D. M., Prigge, M. F., Thenoz, M., Dillman, A. A., Mandler, M. D., et al. (2019). TET-catalyzed 5-carboxylcytosine promotes CTCF binding to suboptimal sequences genome-wide. iScience., 19, 326–339.
Fujii, T., Khawaja, M. R., DiNardo, C. D., Atkins, J. T., & Janku, F. (2016). Targeting isocitrate dehydrogenase (IDH) in cancer. Discovery Medicine, 21(117), 373–380.
Flavahan, W. A., Drier, Y., Liau, B. B., Gillespie, S. M., Venteicher, A. S., Stemmer-Rachamimov, A. O., et al. (2016). Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature., 529(7584), 110–114.
Ito, K., Lee, J., Chrysanthou, S., Zhao, Y., Josephs, K., Sato, H., et al. (2019). Non-catalytic roles of Tet2 are essential to regulate hematopoietic stem and progenitor cell homeostasis. Cell Reports, 28(10), 2480–2490.e4.
Montagner, S., Leoni, C., Emming, S., Della Chiara, G., Balestrieri, C., Barozzi, I., et al. (2016). TET2 regulates mast cell differentiation and proliferation through catalytic and non-catalytic activities. Cell Reports, 15(7), 1566–1579.
Zhang, Q., Zhao, K., Shen, Q., Han, Y., Gu, Y., Li, X., et al. (2015). Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature., 525(7569), 389–393.
Chen, Q., Chen, Y., Bian, C., Fujiki, R., & Yu, X. (2013). TET2 promotes histone O-GlcNAcylation during gene transcription. Nature., 493(7433), 561–564.
Vella, P., Scelfo, A., Jammula, S., Chiacchiera, F., Williams, K., Cuomo, A., et al. (2013). Tet proteins connect the O-linked N-acetylglucosamine transferase Ogt to chromatin in embryonic stem cells. Molecular Cell, 49(4), 645–656.
Montalbán-Loro, R., Lozano-Ureña, A., Ito, M., Krueger, C., Reik, W., Ferguson-Smith, A. C., et al. (2019). TET3 prevents terminal differentiation of adult NSCs by a non-catalytic action at Snrpn. Nature Communications, 10(1), 1–14.
Mi, Y., Gao, X., Dai, J., Ma, Y., Xu, L., & Jin, W. A. (2015). novel function of TET2 in CNS: Sustaining neuronal survival. International Journal of Molecular Sciences, 16(9), 21846–21857.
Huang, Y., Wang, G., Liang, Z., Yang, Y., Cui, L., & Liu, C.-Y. (2016). Loss of nuclear localization of TET2 in colorectal cancer. Clinical Epigenetics, 8, 9.
Yan, H., Tan, L., Liu, Y., Huang, N., Cang, J., & Wang, H. (2020). Ten-eleven translocation methyl-cytosine dioxygenase 2 deficiency exacerbates renal ischemia-reperfusion injury. Clinical Epigenetics, 12(1), 98.
Li, R., Zhou, Y., Cao, Z., Liu, L., Wang, J., Chen, Z., et al. (2017). TET2 loss dysregulates the behavior of bone marrow mesenchymal stromal cells and accelerates Tet2−/−-driven myeloid malignancy progression. Stem Cell Reports, 10(1), 166–179.
Chu, Y., Zhao, Z., Sant, D. W., Zhu, G., Greenblatt, S. M., Liu, L., et al. (2018). Tet2 regulates osteoclast differentiation by interacting with Runx1 and maintaining genomic 5-hydroxymethylcytosine (5hmC). Genomics, Proteomics & Bioinformatics, 16(3), 172–186.
Kamdar, S., Isserlin, R., Van der Kwast, T., & Zlotta, R. A. (2019). Exploring targets of TET2-mediated methylation reprogramming as potential discriminators of prostate cancer progression. Clinical Epigenetics, 11(1), 54.
Wang H, Liu L, Gou M, Huang G, Tian C, Yang J, et al. (2020) Roles of Tet2 in meiosis, fertility and reproductive aging. Protein Cell [Internet]. [cited 2020 Dec 10]; Available from: https://doi.org/10.1007/s13238-020-00805-8.
Nair, V. S., & Oh, K. I. (2014). Down-regulation of Tet2 prevents TSDR demethylation in IL2 deficient regulatory T cells. Biochemical and Biophysical Research Communications, 450(1), 918–924.
Wu, Y., Guo, Z., Liu, Y., Tang, B., Wang, Y., Yang, L., et al. (2013). Oct4 and the small molecule inhibitor, SC1, regulates Tet2 expression in mouse embryonic stem cells. Molecular Biology Reports, 40(4), 2897–2906.
Fischer, A. P., & Miles, S. L. (2017). Silencing HIF-1α induces TET2 expression and augments ascorbic acid induced 5-hydroxymethylation of DNA in human metastatic melanoma cells. Biochemical and Biophysical Research Communications, 490(2), 176–181.
Cheng, J., Guo, S., Chen, S., Mastriano, S. J., Liu, C., D’Alessio, A. C., et al. (2013). An Extensive Network of TET2-Targeting MicroRNAs Regulates Malignant Hematopoiesis. Cell Reports, 5(2), 471–481.
Ren, S., & Xu, Y. (2019). AC016405.3, a novel long noncoding RNA, acts as a tumor suppressor through modulation of TET2 by microRNA-19a-5p sponging in glioblastoma. Cancer Science, 110(5), 1621–1632.
Song, S. J., Ito, K., Ala, U., Kats, L., Webster, K., Sun, S. M., et al. (2013). The oncogenic microRNA miR-22 targets the TET2 tumor suppressor to promote hematopoietic stem cell self-renewal and transformation. Cell Stem Cell, 13(1), 87–101.
Stefan-Lifshitz, M., Karakose, E., Cui, L., Ettela, A., Yi, Z., Zhang, W., et al. (2019). Epigenetic modulation of β cells by interferon-α via PNPT1/mir-26a/TET2 triggers autoimmune diabetes. JCI Insight., 4(5).
Zhaolin, Z., Jiaojiao, C., Peng, W., Yami, L., Tingting, Z., Jun, T., et al. (2019). OxLDL induces vascular endothelial cell pyroptosis through miR-125a-5p/TET2 pathway. Journal of Cellular Physiology, 234(5), 7475–7491.
Bauer, C., Göbel, K., Nagaraj, N., Colantuoni, C., Wang, M., Müller, U., et al. (2015). Phosphorylation of TET proteins is regulated via O-GlcNAcylation by the O-linked N-acetylglucosamine transferase (OGT). The Journal of Biological Chemistry, 290(8), 4801–4812.
Chen, H., Yu, D., Fang, R., Rabidou, K., Wu, D., Hu, D., et al. (2019). TET2 stabilization by 14-3-3 binding to the phosphorylated Serine 99 is deregulated by mutations in cancer. Cell Research, 29(3), 248–250.
Wu, D., Hu, D., Chen, H., Shi, G., Fetahu, I. S., Wu, F., et al. (2018). Glucose-regulated phosphorylation of TET2 by AMPK reveals a pathway linking diabetes to cancer. Nature., 559(7715), 637–641.
Zhang, T., Guan, X., Choi, U. L., Dong, Q., Lam, M. M. T., Zeng, J., et al. (2019). Phosphorylation of TET2 by AMPK is indispensable in myogenic differentiation. Epigenetics & Chromatin, 12(1), 32.
Gwinn, D. M., Shackelford, D. B., Egan, D. F., Mihaylova, M. M., Mery, A., Vasquez, D. S., et al. (2008). AMPK phosphorylation of raptor mediates a metabolic checkpoint. Molecular Cell, 30(2), 214–226.
Hardie, D. G., Ross, F. A., & Hawley, S. A. (2012). AMPK: A nutrient and energy sensor that maintains energy homeostasis. Nature Reviews. Molecular Cell Biology, 13(4), 251–262.
Jeong, J. J., Gu, X., Nie, J., Sundaravel, S., Liu, H., Kuo, W.-L., et al. (2019). Cytokine-regulated phosphorylation and activation of TET2 by JAK2 in hematopoiesis. Cancer Discovery, 9(6), 778–795.
Swamy, M., Pathak, S., Grzes, K. M., Damerow, S., Sinclair, L. V., van Aalten, D. M. F., et al. (2016). Glucose and glutamine fuel protein O-GlcNAcylation to control T cell self-renewal and malignancy. Nature Immunology, 17(6), 712–720.
Ramanathan, H. N., & Ye, Y. (2012). Cellular strategies for making monoubiquitin signals. Critical Reviews in Biochemistry and Molecular Biology, 47(1), 17–28.
Nakagawa, T., Lv, L., Nakagawa, M., Yu, Y., Yu, C., D’Alessio, A. C., et al. (2015). CRL4VprBP E3 ligase promotes monoubiquitylation and chromatin binding of TET dioxygenases. Molecular Cell, 57(2), 247–260.
Kosmider, O., Gelsi-Boyer, V., Cheok, M., Grabar, S., Della-Valle, V., Picard, F., et al. (2009). TET2 mutation is an independent favorable prognostic factor in myelodysplastic syndromes (MDSs). Blood., 114(15), 3285–3291.
Delhommeau, F., Dupont, S., Della Valle, V., James, C., Trannoy, S., Massé, A., et al. (2009). Mutation in TET2 in myeloid cancers. The New England Journal of Medicine, 360(22), 2289–2301.
Zhang, Y. W., Wang, Z., Xie, W., Cai, Y., Xia, L., Easwaran, H., et al. (2017). Acetylation enhances TET2 function in protecting against abnormal DNA methylation during oxidative stress. Molecular Cell, 65(2), 323–335.
Yin, R., Mao, S.-Q., Zhao, B., Chong, Z., Yang, Y., Zhao, C., et al. (2013). Ascorbic acid enhances Tet-mediated 5-methylcytosine oxidation and promotes DNA demethylation in mammals. Journal of the American Chemical Society, 135(28), 10396–10403.
Unruh, D., Zewde, M., Buss, A., Drumm, M. R., Tran, A. N., Scholtens, D. M., et al. (2019). Methylation and transcription patterns are distinct in IDH mutant gliomas compared to other IDH mutant cancers. Scientific Reports, 9(1), 8946.
Figueroa, M. E., Abdel-Wahab, O., Lu, C., Ward, P. S., Patel, J., Shih, A., et al. (2010). Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell, 18(6), 553–567.
Xu, W., Yang, H., Liu, Y., Yang, Y., Wang, P., Kim, S.-H., et al. (2011). Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell, 19(1), 17–30.
Ward, P. S., Cross, J. R., Lu, C., Weigert, O., Abel-Wahab, O., Levine, R. L., et al. (2012). Identification of additional IDH mutations associated with oncometabolite R (−)-2-hydroxyglutarate production. Oncogene., 31(19), 2491–2498.
Guan, Y., Tiwari, A. D., Phillips, J. G., Hasipek, M., Grabowski, D., Kerr, C. M., et al. (2019). TET dioxygenase inhibition as a therapeutic strategy in TET2 mutant myeloid neoplasia. Blood., 134(Supplement_1), 880–880.
Itzykson, R., Kosmider, O., Renneville, A., Morabito, M., Berthon, C., Ades, L., et al. (2012). Comprehensive genetic screening of chronic myelomonocytic leukemias (CMML). Blood., 120(21), 3811–3811.
Kao, H.-W., Kuo, M.-C., Wang, P.-N., Wu, J.-H., Lin, T.-H., Shih, Y.-S., et al. (2015). Clonal evolution of gene mutations involving DNA methylation in the progression of CMML to secondary AML. Blood., 126(23), 4120–4120.
Kosmider, O., LaRochelle, O., Coude, M.-M., Mas, V. M.-D., Delabesse, E., Cornillet-Lefebvre, P., et al. (2011). IDH1/2, TET2 and DNMT3A mutations are not mutually exclusive in secondary acute myeloid leukemias. Blood., 118(21), 3556–3556.
Chang, J., Xie, M., Shah, V. R., Schneider, M. D., Entman, M. L., Wei, L., et al. (2006). Activation of Rho-associated coiled-coil protein kinase 1 (ROCK-1) by caspase-3 cleavage plays an essential role in cardiac myocyte apoptosis. Proceedings of the National Academy of Sciences, 103(39), 14495–14500.
Chen, J., Wu, Y., Zhang, L., Fang, X., & Hu, X. (2019). Evidence for calpains in cancer metastasis. Journal of Cellular Physiology, 234(6), 8233–8240.
Wang, Y., & Zhang, Y. (2014). Regulation of TET protein stability by calpains. Cell Reports, 6(2), 278–284.
Sardina, J. L., Collombet, S., Tian, T. V., Gómez, A., Di Stefano, B., Berenguer, C., et al. (2018). Transcription factors drive Tet2-mediated enhancer demethylation to reprogram cell fate. Cell Stem Cell, 23(5), 727–741.e9.
Guilhamon, P., Eskandarpour, M., Halai, D., Wilson, G. A., Feber, A., Teschendorff, A. E., et al. (2013). Meta-analysis of IDH-mutant cancers identifies EBF1 as an interaction partner for TET2. Nature Communications, 4, 2166.
Wang, Y., Xiao, M., Chen, X., Chen, L., Xu, Y., Lv, L., et al. (2015). WT1 recruits TET2 to regulate its target gene expression and suppress leukemia cell proliferation. Molecular Cell, 57(4), 662–673.
Yamamoto, M., Yamazaki, S., Uematsu, S., Sato, S., Hemmi, H., Hoshino, K., et al. (2004). Regulation of Toll/IL-1-receptor-mediated gene expression by the inducible nuclear protein IκBζ. Nature., 430(6996), 218–222.
Treiber, T., Mandel, E. M., Pott, S., Györy, I., Firner, S., Liu, E. T., et al. (2010). Early B cell factor 1 regulates B cell gene networks by activation, repression, and transcription-independent poising of chromatin. Immunity., 32(5), 714–725.
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2019). STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Research, 47(D1), D607–D613.
Warde-Farley, D., Donaldson, S. L., Comes, O., Zuberi, K., Badrawi, R., Chao, P., et al. (2010). The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Research, 38(Web Server issue), W214–W220.
Weissmann, S., Alpermann, T., Grossmann, V., Kowarsch, A., Nadarajah, N., Eder, C., et al. (2012). Landscape of TET2 mutations in acute myeloid leukemia. Leukemia., 26(5), 934–942.
Ko, M., Huang, Y., Jankowska, A. M., Pape, U. J., Tahiliani, M., Bandukwala, H. S., et al. (2010). Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature., 468(7325), 839–843.
Musialik, E., Bujko, M., Wypych, A., Matysiak, M., & Siedlecki, J. A. (2014). TET2 promoter DNA methylation and expression analysis in pediatric B-cell acute lymphoblastic leukemia. Hematology Reports, 6(1), 5333.
Roche-Lestienne, C., Alice, M., Labis, E., Nibourel, O., Coiteux, V., Guilhot, J., et al. (2010). Mutations of TET2, IDH1, IDH2 and ASXL1 in chronic myeloid leukemia. Blood., 116(21), 3377–3377.
Song, F., Amos, C. I., Lee, J. E., Lian, C. G., Fang, S., Liu, H., et al. (2014). Identification of a melanoma susceptibility locus and somatic mutation in TET2. Carcinogenesis., 35(9), 2097–2101.
Nickerson, M. L., Das, S., Im, K. M., Turan, S., Berndt, S. I., Li, H., et al. (2017). TET2 binds the androgen receptor and Loss is associated with prostate cancer. Oncogene., 36(15), 2172–2183.
Itoh, H., Kadomatsu, T., Tanoue, H., Yugami, M., Miyata, K., Endo, M., et al. (2018). TET2-dependent IL-6 induction mediated by the tumor microenvironment promotes tumor metastasis in osteosarcoma. Oncogene., 37(22), 2903–2920.
Sajadian, S. O., Ehnert, S., Vakilian, H., Koutsouraki, E., Damm, G., Seehofer, D., et al. (2015). Induction of active demethylation and 5hmC formation by 5-azacytidine is TET2 dependent and suggests new treatment strategies against hepatocellular carcinoma. Clinical Epigenetics, 7, 98.
Zhou, K., Guo, H., Zhang, J., Zhao, D., Zhou, Y., Zheng, Z., et al. (2019). Potential role of TET2 in gastric cancer cisplatin resistance. Pathology, Research and Practice, 215(11), 152637.
Schoeler, K., Aufschnaiter, A., Messner, S., Derudder, E., Herzog, S., Villunger, A., et al. (2019). TET enzymes control antibody production and shape the mutational landscape in germinal centre B cells. The FEBS Journal, 286(18), 3566–3581.
Dominguez, P. M., Ghamlouch, H., Rosikiewicz, W., Kumar, P., Béguelin, W., Fontán, L., et al. (2018). TET2 deficiency causes germinal center hyperplasia, impairs plasma cell differentiation, and promotes B-cell lymphomagenesis. Cancer Discovery, 8(12), 1632–1653.
Zhao, Z., Chen, L., Dawlaty, M. M., Pan, F., Weeks, O., Zhou, Y., et al. (2015). Combined loss of Tet1 and Tet2 promotes B cell, but not myeloid malignancies, in mice. Cell Reports, 13(8), 1692–1704.
Tanaka, S., Ise, W., Inoue, T., Ito, A., Ono, C., Shima, Y., et al. (2020). Tet2 and Tet3 in B cells are required to repress CD86 and prevent autoimmunity. Nature Immunology, 21(8), 950–961.
Kamdar, S., Isserlin, R., Van der Kwast, T., Zlotta, A. R., Bader, G. D., Fleshner, N. E., et al. (2019). Exploring targets of TET2-mediated methylation reprogramming as potential discriminators of prostate cancer progression. Clinical Epigenetics, 11(1), 54.
Xu, Y.-P., Lv, L., Liu, Y., Smith, M. D., Li, W.-C., Tan, X.-M., et al. (2019). Tumor suppressor TET2 promotes cancer immunity and immunotherapy efficacy. The Journal of Clinical Investigation, 130, 4316–4331.
Bonvin, E., Radaelli, E., Bizet, M., Luciani, F., Calonne, E., Putmans, P., et al. (2019). TET2-dependent hydroxymethylome plasticity reduces melanoma initiation and progression. Cancer Research, 79(3), 482–494.
Zhang, L.-Y., Li, P.-L., Wang, T.-Z., & Zhang, X.-C. (2015). Prognostic values of 5-hmC, 5-mC and TET2 in epithelial ovarian cancer. Archives of Gynecology and Obstetrics, 292(4), 891–897.
Peng, J., Yang, Q., Li, A.-F., Li, R.-Q., Wang, Z., Liu, L.-S., et al. (2016). Tet methylcytosine dioxygenase 2 inhibits atherosclerosis via upregulation of autophagy in ApoE-/- mice. Oncotarget, 7(47), 76423–76436.
Lee, M., Bryan, K., Peipei, L., Elizabeth, E., Katie, L., Wei, C., et al. (2019). Epigenomic analysis of Parkinson’s disease neurons identifies Tet2 loss as neuroprotective. bioRxiv, 779785.
Roca, F. J., Loomans, H. A., Wittman, A. T., Creighton, C. J., & Hawkins, S. M. (2016). Ten-eleven translocation genes are downregulated in endometriosis. Current Molecular Medicine, 16(3), 288–298.
Ichiyama, K., Chen, T., Wang, X., Yan, X., Kim, B.-S., Tanaka, S., et al. (2015). The methylcytosine dioxygenase Tet2 promotes DNA demethylation and activation of cytokine gene expression in T cells. Immunity., 42(4), 613–626.
Yue, X., Lio, C.-W. J., Samaniego-Castruita, D., Li, X., & Rao, A. (2019). Loss of TET2 and TET3 in regulatory T cells unleashes effector function. Nature Communications, 10(1), 2011.
Braun, T., & Fenaux, P. (2013). Myelodysplastic syndromes (MDS) and autoimmune disorders (AD): Cause or consequence? Best Practice & Research. Clinical Haematology, 26(4), 327–336.
Komrokji, R. S., Kulasekararaj, A., Al Ali, N. H., Kordasti, S., Bart-Smith, E., Craig, B. M., et al. (2016). Autoimmune diseases and myelodysplastic syndromes. American Journal of Hematology, 91(5), E280–E283.
Oh, Y.-J., Shin, D.-Y., Hwang, S. M., Kim, S.-M., Im, K., Park, H. S., et al. (2020). Mutation of ten-eleven translocation-2 is associated with increased risk of autoimmune disease in patients with myelodysplastic syndrome. The Korean Journal of Internal Medicine, 35(2), 457–464.
de Andres, M. C., Perez-Pampin, E., Calaza, M., Santaclara, F. J., Ortea, I., Gomez-Reino, J. J., et al. (2015). Assessment of global DNA methylation in peripheral blood cell subpopulations of early rheumatoid arthritis before and after methotrexate. Arthritis Research & Therapy, 17, 233.
Li, X., Xiong, X., & Yi, C. (2017). Epitranscriptome sequencing technologies: Decoding RNA modifications. Nature Methods, 14(1), 23–31.
Huber, S. M., van Delft, P., Mendil, L., Bachman, M., Smollett, K., Werner, F., et al. (2015). Formation and abundance of 5-hydroxymethylcytosine in RNA. Chembiochem., 16(5), 752–755.
Squires, J. E., Patel, H. R., Nousch, M., Sibbritt, T., Humphreys, D. T., Parker, B. J., et al. (2012). Widespread occurrence of 5-methylcytosine in human coding and noncoding RNA. Nucleic Acids Research, 40(11), 5023–5033.
Basanta-Sanchez, M., Wang, R., Liu, Z., Ye, X., Li, M., Shi, X., et al. (2017). TET1-mediated oxidation of 5-formylcytosine (5fC) to 5-carboxycytosine (5caC) in RNA. Chembiochem : a European journal of chemical biology, 18(1), 72–76.
Yang, X., Yang, Y., Sun, B.-F., Chen, Y.-S., Xu, J.-W., Lai, W.-Y., et al. (2017). 5-methylcytosine promotes mRNA export-NSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Research, 27(5), 606–625.
Amort, T., Rieder, D., Wille, A., Khokhlova-Cubberley, D., Riml, C., Trixl, L., et al. (2017). Distinct 5-methylcytosine profiles in poly(A) RNA from mouse embryonic stem cells and brain. Genome Biology, 18(1), 1.
Amort, T., Soulière, M. F., Wille, A., Jia, X.-Y., Fiegl, H., Wörle, H., et al. (2013). Long noncoding RNAs as targets for cytosine methylation. RNA Biology, 10(6), 1002–1008.
Amort, M., Nachbauer, B., Tuzlak, S., Kieser, A., Schepers, A., Villunger, A., et al. (2015). Expression of the vault RNA protects cells from undergoing apoptosis. Nature Communications, 6, 7030.
Hussain, S., Sajini, A. A., Blanco, S., Dietmann, S., Lombard, P., Sugimoto, Y., et al. (2013). NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Reports, 4(2), 255–261.
Persson, H., Kvist, A., Vallon-Christersson, J., Medstrand, P., Borg, A., & Rovira, C. (2009). The noncoding RNA of the multidrug resistance-linked vault particle encodes multiple regulatory small RNAs. Nature Cell Biology, 11(10), 1268–1271.
Sajini, A. A., Choudhury, N. R., Wagner, R. E., Bornelöv, S., Selmi, T., Spanos, C., et al. (2019). Loss of 5-methylcytosine alters the biogenesis of vault-derived small RNAs to coordinate epidermal differentiation. Nat Commun., 10(1), 11, 2550.
Zhou, Z., Luo, M., Straesser, K., Katahira, J., Hurt, E., & Reed, R. (2000). The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature., 407(6802), 401–405.
Fu, L., Guerrero, C. R., Zhong, N., Amato, N. J., Liu, Y., Liu, S., et al. (2014). Tet-mediated formation of 5-hydroxymethylcytosine in RNA. Journal of the American Chemical Society, 136(33), 11582–11585.
Delatte, B., Wang, F., Ngoc, L. V., Collignon, E., Bonvin, E., Deplus, R., et al. (2016). Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science., 351(6270), 282–285.
DeNizio, J. E., Liu, M. Y., Leddin, E. M., Cisneros, G. A., & Kohli, R. M. (2019). Selectivity and promiscuity in TET-mediated oxidation of 5-methylcytosine in DNA and RNA. Biochemistry., 58(5), 411–421.
Leddin, E. M., & Cisneros, G. A. (2019). Comparison of DNA and RNA substrate effects on TET2 structure. Advances in Protein Chemistry and Structural Biology, 117, 91–112.
Haag, S., Sloan, K. E., Ranjan, N., Warda, A. S., Kretschmer, J., Blessing, C., et al. (2016). NSUN3 and ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation. The EMBO Journal, 35(19), 2104–2119.
Kawarada, L., Suzuki, T., Ohira, T., Hirata, S., Miyauchi, K., & Suzuki, T. (2017). ALKBH1 is an RNA dioxygenase responsible for cytoplasmic and mitochondrial tRNA modifications. Nucleic Acids Research, 45(12), 7401–7415.
Huber, S. M., van Delft, P., Tanpure, A., Miska, E. A., & Balasubramanian, S. (2017). 2′-O-Methyl-5-hydroxymethylcytidine: A second oxidative derivative of 5-methylcytidine in RNA. Journal of the American Chemical Society, 139(5), 1766–1769.
Zhang, Y.-F., Qi, C.-B., Yuan, B.-F., & Feng, Y.-Q. (2019). Determination of cytidine modifications in human urine by liquid chromatography-mass spectrometry analysis. Analytica Chimica Acta, 1081, 103–111.
Gaj, T., Sirk, S. J., Shui, S.-L., & Liu, J. (2016). Genome-editing technologies: Principles and applications. Cold Spring Harbor Perspectives in Biology, 8(12).
Sano, S., Oshima, K., Wang, Y., Katanasaka, Y., Sano, M., & Walsh, K. (2018). CRISPR-mediated gene editing to assess the roles of Tet2 and Dnmt3a in clonal hematopoiesis and cardiovascular disease. Circulation Research, 123(3), 335–341.
Yu, M., Han, D., Hon, G. C., & He, C. (2018). Tet-assisted bisulfite sequencing (TAB-seq). Methods in Molecular Biology, Clifton NJ., 1708, 645–663.
Yu, M., Hon, G. C., Szulwach, K. E., Song, C.-X., Jin, P., Ren, B., et al. (2012). Tet-assisted bisulfite sequencing of 5-hydroxymethylcytosine. Nature Protocols, 7(12), 2159–2170.
Yu, M., Hon, G. C., Szulwach, K. E., Song, C.-X., Zhang, L., Kim, A., et al. (2012). Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell., 149(6), 1368–1380.
Skvortsova, K., Zotenko, E., Luu, P.-L., Gould, C. M., Nair, S. S., Clark, S. J., et al. (2017). Comprehensive evaluation of genome-wide 5-hydroxymethylcytosine profiling approaches in human DNA. Epigenetics & Chromatin, 10(1), 16.
Jin, S.-G., Kadam, S., & Pfeifer, G. P. (2010). Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Research, 38(11), e125.
Booth, M. J., Branco, M. R., Ficz, G., Oxley, D., Krueger, F., Reik, W., et al. (2012). Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science., 336(6083), 934–937.
Song, C.-X., Szulwach, K. E., Fu, Y., Dai, Q., Yi, C., Li, X., et al. (2011). Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nature Biotechnology, 29(1), 68–72.
Rostovtsev, V. V., Green, L. G., Fokin, V. V., & Sharpless, K. B. (2002). A stepwise huisgen cycloaddition process: Copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angewandte Chemie (International Ed. in English), 41(14), 2596–2599.
Nestor, C. E., & Meehan, R. R. (2014). Hydroxymethylated DNA immunoprecipitation (hmeDIP). Methods in Molecular Biology, Clifton NJ., 1094, 259–267.
Shenoy, N., Bhagat, T., Nieves, E., Stenson, M., Lawson, J., Choudhary, G. S., et al. (2017). Upregulation of TET activity with ascorbic acid induces epigenetic modulation of lymphoma cells. Blood Cancer Journal, 7(7), e587–e587.
Davis, T., & Vaisvila, R. (2011). High sensitivity 5-hydroxymethylcytosine detection in Balb/C brain tissue. Journal of Visualized Experiments, 48.
Zahid, O. K., Zhao, B. S., He, C., & Hall, A. R. (2016). Quantifying mammalian genomic DNA hydroxymethylcytosine content using solid-state nanopores. Scientific Reports, 6, 29565.
Song, C.-X., Clark, T. A., Lu, X.-Y., Kislyuk, A., Dai, Q., Turner, S. W., et al. (2011). Sensitive and specific single-molecule sequencing of 5-hydroxymethylcytosine. Nature Methods, 9(1), 75–77.
Acknowledgements
We would like to express our thanks to the Department of Biochemistry, All India Institute of Medical Science, New Delhi, for providing logistics and space.
Funding
SK received funding from AIIMS - New Delhi, and SERB - Department of Science and Technology, Government of India.
Author information
Authors and Affiliations
Contributions
AS and KP drafted the manuscript. IC and RS assisted in writing. AC helped in the critical discussion of the manuscript. RD and SK shared the idea and oversaw the whole work.
Corresponding authors
Ethics declarations
Ethical Approval
Ethical approval is not required since this is a review article.
Consent to Participate
N/A as no human subjects were enrolled in this study.
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Seethy, A., Pethusamy, K., Chattopadhyay, I. et al. TETology: Epigenetic Mastermind in Action. Appl Biochem Biotechnol 193, 1701–1726 (2021). https://doi.org/10.1007/s12010-021-03537-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-021-03537-5