Abstract
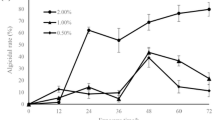
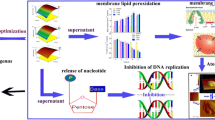
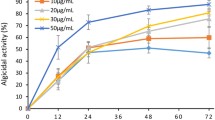
The purpose of this study was to examine the efficacy of the algicidal bacterium Sagittula stellata on the cell lysis of Nannochloropsis oceanica, a microalga found in the marine environment, in order to extract intracellular valuables. Algicidal bacteria are capable of lysing algal cell walls while keeping lipids and proteins intact yet separated. We obtained these microbes from locations with consistent algae blooms and found that the bacterium Sagittula stellata displayed significant algicidal properties toward Nannochloropsis oceanica, achieving an algicidal rate of 80.1%. We detected a decrease of 66.2% in in vivo fluorescence intensity in algae cultures, obtained a recoverable crude lipid content of 23.3% and a polyunsaturated fatty acid (PUFA) ratio of 29.0% of bacteria-treated algae, and observed the lysis of the cell membrane and the structure of the nucleus of algae. We also identified the inhibited transcription of the ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit (rbcS) gene and proliferating cell nuclear antigen (PCNA)–related genes and the upregulated heat shock protein (hsp) gene in algal cells during bacterial exposure. Our results indicate that Sagittula stellata effectively lysed microalgae cells, allowing the recovery of intracellular valuables. The algicidal method of Sagittula stellata on Nannochloropsis oceanica cells was confirmed to be a direct attack (or predation), followed by an indirect attack through the secretion of extracellular algicidal compounds. This study provides an important framework for the broad application of algicidal microorganisms in algal cell disruption and the production of intracellular valuables.
Graphical Abstract







Similar content being viewed by others
Data Availability
The data sets supporting the results of this article are included within the article.
Abbreviations
- ARA:
-
Arachidonic acid
- BS:
-
Bacterial supernatant
- CAFD:
-
Chlorophyll a fluorescence intensity
- CFU:
-
Colony-forming units
- DHA:
-
Docosahexaenoic acid
- EPA:
-
Eicosapentaenoic acid
- FAMEs:
-
Fatty acid methyl esters
- GC:
-
Gas chromatography
- hsp :
-
Heat shock protein
- MMBM:
-
Modified Marine Broth 2216 medium
- MUFAs:
-
Monounsaturated fatty acids
- N. oceanica :
-
Nannochloropsis oceanica
- PUFAs:
-
Polyunsaturated fatty acids
- PCNA:
-
Proliferating cell nuclear antigen
- rbc :
-
S: Ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit
- S. stellata :
-
Sagittula stellata
- SFAs:
-
Saturated fatty acids
- TEM:
-
Transmission electron microscopy
References
Manage, P. M., Kawabata, Z., & Nakano, S. (2000). Algicidal effect of the bacterium Alcaligenes denitrificans on Microcystisspp. Aquatic Microbial Ecology, 22, 111–117.
Natrah, F. M. I., Bossier, P., Sorgeloos, P., Yusoff, F. M., Defoirdt, T. (2014). Significance of microalgal–bacterial interactions for aquaculture. Reviews in Aquaculture, 648–661.
Zohdi, E., & Abbaspour, M. (2019). Harmful algal blooms (red tide): a review of causes, impacts and approaches to monitoring and prediction. International Journal of Environmental Sciences, 16(3), 1789–1806.
Roth, P. B., Twiner, M. J., Mikulski, C. M., Barnhorst, A. B., & Doucette, G. J. (2008). Comparative analysis of two algicidal bacteria active against the red tide dinoflagellate Karenia brevis. Harmful Algae, 7, 682–691.
Meyer, N., Bigalke, A., Kaulfuß, A., & Pohnert, G. (2017). Strategies and ecological roles of algicidal bacteria. FEMS Microbiology Reviews, 41(6), 880–899.
Anderson, D. M., Cembella, A. D., & Hallegraeff, G. M. (2012). Progress in understanding harmful algal blooms: paradigm shifts and new technologies for research, monitoring, and management. Annual Review of Marine Science, 4(1), 143–176.
Li, Z., Geng, M., Yang, H. (2015). Algicidal activity of Bacillus sp. lzh-5 and its algicidal compounds against Microcystis aeruginosa. Applied Microbiology and Biotechnology, 99(2), 981–990.
van Tol, H. M., Amin, S. A., & Armbrust, E. V. (2017). Ubiquitous marine bacterium inhibits diatom cell division. ISME Journal, 11, 31–42.
Stewart, J. R., & Brown, R. M. (1969). Cytophaga that kills or lyses algae. Science, New Series, 164(3887), 1523–1524.
Zhang, H., Yu, Z. L., Huang, Q., Xiao, X., Wang, X., Zhang, F., et al. (2011). Isolation, identification and characterization of phytoplankton-lytic bacterium CH-22 against Microcystis aeruginosa. Limnologica, 41, 70–77.
Pokrzywinski, K. L., Tilney, C. L., Modla, S., Caplan, J. L., Ross, J., Warner, M. E., et al. (2017). Effects of the bacterial algicide IRI-160AA on cellular morphology of harmful dinoflagellates. Harmful Algae, 62, 127–135.
Yu, X., Cai, G., Wang, H., Hu, Z., Zheng, W., Lei, X., et al. (2018). Fast-growing algicidalStreptomyces sp. U3 and its potential in harmful algal bloom controls. Journal of Hazardous Materials, 341, 138–149.
Deshmukh, S., Bala, K., & Kumar, R. (2019). Selection of microalgae species based on their lipid content, fatty acid profile and apparent fuel properties for biodiesel production. Environmental Science and Pollution Research, 26(1), 24462–24473.
Chen, Y., Xu, X., & Vaidyanathan, S. (2020). Influence of gas management on biochemical conversion of CO2 by microalgae for biofuel production. Applied Energy, 261, 114420.
Reis, A., & Gouveia, L. (2016). Low cost microalgal production for biofuels: a review. Current Biotechnology, 5(4), 266–276.
Menegazzo, M. L., & Fonseca, G. G. (2019). Biomass recovery and lipid extraction processes for microalgae biofuels production: a review. Renewable and Sustainable Energy Reviews, 107, 87–107.
Kanda, H., Hoshino, R., Murakami, K., Wahyudiono, Z., & Q., Goto, M. . (2020). Lipid extraction from microalgae covered with biomineralized cell walls using liquefied dimethyl ether. Fuel, 262, 116590.
Show, K. Y., Lee, D. J., Tay, J. H., Lee, T. M., & Chang, J. S. (2015). Microalgal drying and cell disruption–recent advances. Bioresource technology, 184, 258–266.
Lee, S. Y., Cho, J. M., Chang, Y. K., & Oh, Y. K. (2017). Cell disruption and lipid extraction for microalgalbiorefineries: a review. Bioresource Technology, 244, 1317–1328.
Wang, M., Cheng, H., Chen, S., Wen, S., Wu, X., & Zhang, D. (2018). Microalgal cell disruption via extrusion for the production of intracellular valuables. Energy, 142, 339–345.
Wang, M., Yuan, W., Jiang, X., Yun, J., & Wang, Z. (2014). Disruption of microalgal cells using high-frequency focused ultrasound. Bioresource Technology, 153(2), 315–321.
Spiden, E. M., Yap, B. H., Hill, D. R. A., Kentish, S. E., Scales, P. J., & Martin, G. J. O. (2013). Quantitative evaluation of the ease of rupture of industrially promising microalgae by high pressure homogenization. Bioresource Technology, 140, 165–171.
Prabakaran, P., & Ravindran, A. D. (2011). A comparative study on effective cell disruption methods for lipid extraction from microalgae. Letters in applied microbiology, 53(2), 150–154.
Enamala, M. K., Enamala, S., Chavali, M., Donepudi, J., Donepudi, R., Kolapalli, B., et al. (2018). Production of biofuels from microalgae - a review on cultivation, harvesting, lipid extraction, and numerous applications of microalgae. Renewable and Sustainable Energy Reviews, 94, 49–68.
Demuez, M., Gonzalez-Fernandez, C., & Ballesteros, M. (2015). Algicidal microorganisms and secreted algicides: new tools to induce microalgal cell disruption. Biotechnology Advances, 33(8), 1615–1625.
Boden, R., Murrel, J. C., & Schäfer, H. (2011). Dimethylsulfide is an energy source for the heterotrophic marine bacterium Sagittula stellata. FEMS Microbiology Letters, 322(2), 188–193.
Ding, Y. X., Chin, W. C., Rodriguez, A., Hung, C. C., Santschi, P. H., & Verdugo, P. (2008). Amphiphilic exopolymers from Sagittula stellata induce DOM self-assembly and formation of marine microgels. Marine Chemistry, 112(1), 11–19.
González, J. M., Mayer, F., Moran, M. A., Hodson, R. E., Whitman, W. B. (1997). Sagittula stellata gen. nov., sp. nov., a lignin-transforming bacterium from a coastal environment. International Journal of Systematic Bacteriology, 47(3), 773–780.
Wang, M., & Yuan, W. (2014). Bacterial lysis of microalgal cells. Journal of Sustainable Bioenergy Systems, 4, 243–248.
Livak, K. J., & Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the2−△△Ct method. Methods, 25(4), 402–408.
Furusawa, G., Yoshikawa, T., Yasuda, A., Sakata, T. (2003). Algicidal activity and gliding motility of Saprospira sp. SS98-5. Canadian Journal of Microbiology, 49(2), 92–100.
Volkman, J. K., Brown, M. R., Dunstan, G. A., & Jeffrey, S. W. (2010). The biochemical composition of marine microalgae from the class Eustigmatophyceae. Journal of Phycology, 29(1), 69–78.
Patil, V., Källqvist, T., Olsen, E., Vogt, G., & Gislerød, H. R. (2007). Fatty acid composition of 12 microalgae for possible use in aquaculture feed. Aquaculture International, 15(1), 1–9.
Melo, M. M. R., Sapatinha, M., Pinheiro, J., Lemos, M. F. L., Bandarra, N. M., Batista, I., et al. (2020). Supercritical CO2 extraction of Aurantiochytrium sp. biomass for the enhanced recovery of omega-3 fatty acids and phenolic compounds. Journal of CO2 Utilization, 38, 24–31.
Hu, X., Yin, P., Zhao, L., & Yu, Q. (2015). Characterization of cell viability in Phaeocystis globose cultures exposed to marine algicidal bacteria. Biotechnology and Bioprocess Engineering, 20(1), 58–66.
Domozych, D. S., & Domozych, C. E. (2014). Multicellularity in green algae: upsizing in a walled complex. Frontiers in plant science, 5, 1–8.
Genkov, T., Meyer, M., Griffiths, H., & Spreitzer, R. J. (2010). Functional hybrid rubisco enzymes with plant small subunits and algal large subunits: engineered rbcS cDNA for expression in Chlamydomonas. Journal of Biological Chemistry, 285(26), 19833–19841.
Liu, F., Sun, X., Wang, W., Liang, Z., & Wang, F. (2014). De novo transcriptome analysis-gained insights into physiological and metabolic characteristics of Sargassum thunbergii (Fucales, Phaeophyceae). Journal of Applied Phycology, 26(3), 1519–1526.
Boehm, E. M., Gildenberg, M. S., & Washington, M. T. (2016). The many roles of PCNA in eukaryotic DNA replication. The Enzymes, 39, 231–254.
Kim, Y. S., Son, H. J., & Jeong, S. Y. (2015). Isolation of an algicide from a marine bacterium and its effects against the toxic dinoflagellate Alexandrium catenella and other harmful algal bloom species. Journal of Microbiology, 53(8), 511–517.
Petropoulos, K., Bodini, S. F., Fabiani, L., Micheli, L., Porchetta, A., Piermarini, S., et al. (2019). Re-modeling ELISA kits embedded in an automated system suitable for on-line detection of algal toxins in seawater. Sensors and Actuators B: Chemical, 283, 865–872.
Shi, X., Liu, L., Li, Y., Xiao, Y., Ding, G., Lin, S., et al. (2018). Isolation of an algicidal bacterium and its effects against the harmful-algal-bloom dinoflagellate prorocentrum donghaiense (dinophyceae). Harmful Algae, 80, 72–79.
Wang, Y., Li, S., Liu, G., Li, X., Yang, Q., Xu, Y., et al. (2020). Continuous production of algicidal compounds against Akashiwo sanguinea via a Vibrio sp. co-culture. Bioresource Technology, 295, 122246.
Cai, G., Yang, X., Lai, Q., Yu, X., Zhang, H., Li, Y., et al. (2016). Lysing bloom-causing alga Phaeocystis globosa with microbial algicide: an efficient process that decreases the toxicity of algal exudates. Scientific Reports, 6(1), 20081.
Lee, A. K., Lewis, D. M., & Ashman, P. J. (2012). Disruption of microalgal cells for theextraction of lipids for biofuels: processes and specific energy requirements. Biomass and Bioenergy, 46, 89–101.
Halim, R., Harun, R., Danqua, M. K., & Webley, P. A. (2012). Microalgal cell disruption for biofuel development. Applied Energy, 91(1), 116–121.
Cheng, J., Huang, R., Li, T., Zhou, J., & Cen, K. (2015). Physicochemical characterization of wet microalgal cells disrupted with instant catapult steam explosion for lipid extraction. Bioresource Technology, 191, 66–72.
Park, J. Y., Nam, B., Choi, S. A., Oh, Y. K., & Lee, J. S. (2014). Effects of anionic surfactant on extraction of free fatty acid from Chlorella vulgaris. Bioresource Technology, 166, 620–624.
Chen, L., Li, R., Ren, X., & Liu, T. (2016). Improved aqueous extraction of microalgal lipid by combined enzymatic and thermal lysis from wet biomass of Nannochloropsis oceanica. Bioresource Technology, 214, 138–143.
Zhang, S., Du, X., Zhu, J., Meng, C., Zhou, J., & Zou, P. (2020). The complete genome sequence of the algicidal bacterium Bacillus subtilis strain JA and the use of quorum sensing to evaluate its antialgal ability. Biotechnology Reports, 25, e00421.
Shi, S. Y., Liu, Y. D., Shen, Y., Li, G., & Li., D. . (2006). Lysis of Aphanizomenon flos-aquae (cyanobacterium) by a bacterium Bacillus cereus. Biological Control, 39(3), 345–351.
Kim, J. D., Kim, J. Y., Park, J. K., & Lee, C. G. (2009). Selective control of the prorocentrum minimum harmful algal blooms by a novel algal-lytic bacterium Pseudoalteromonas haloplanktis AFMB-008041. Marine Biotechnology, 11(4), 463–472.
Funding
This research was financially supported by the National Natural Science Foundation of China (41877387). We thank Dr. Mary Ann Moran and Ms. Christa Smith at University of Georgia for providing the bacteria and assisting with its cultivation.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by Lifu Wang, Shuwen Zhao, and Shanshan Li. The first draft of the manuscript was written by Meng Wang and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no competing interests.
Ethical Approval
Not applicable
Consent to Participate
Not applicable
Consent to Publish
Not applicable
Rights and permissions
About this article
Cite this article
Wang, M., Yuan, W.q., Chen, S. et al. Algal Lysis by Sagittula stellata for the Production of Intracellular Valuables. Appl Biochem Biotechnol 193, 2516–2533 (2021). https://doi.org/10.1007/s12010-021-03502-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-021-03502-2