Abstract
The unprecedented coronavirus SARS-CoV-2 outbreak at Wuhan, China, caused acute respiratory infection to humans. There is no precise vaccine/therapeutic agents available to combat the COVID-19 disease. Some repurposed drugs are saving the life of diseased, but the complete cure is relatively less. Several drug targets have been reported to inhibit the SARS-CoV-2 virus infection, in that TMPRSS2 (transmembrane protease serine 2) is one of the potential targets; inhibiting this protease stops the virus entry into the host human cell. Camostat mesylate, nafamostat, and leupeptin are the drugs, in which the first two drugs are being used for COVID-19 and leupeptin also tested. To consider these drugs as the repurposed drug for COVID-19, it is essential to understand their binding affinity and stability with TMPRSS2. In the present study, we performed the molecular docking and molecular dynamics (MD) simulation of these molecules with the TMPRSS2. The docking study reveals that leupeptin molecule strongly binds with TMPRSS2 protein than the other two drug molecules. The RMSD and RMSF values of MD simulation confirm that leupeptin and the amino acids of TMPRSS2 are very stable than the other two molecules. Furthermore, leupeptin forms interactions with the key amino acids of TMPRSS2 and the same have been maintained during the MD simulations. This structural and dynamical information is useful to evaluate these drugs to be used as repurposed drugs, however, the strong binding profile of leupeptin with TMPRSS2, suggests, it may be considered as a repurposed drug for COVID-19 disease after clinical trial.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
For the past three decades, the outbreak of new viruses causes strange epidemic diseases and high mortality; among these, the influenza, hepatitis, and HIV viruses are highly replicative in nature and no drugs are available neither to stop the infection nor to give the complete cure. Coronavirus is an RNA virus, which was emerged and evolved to different types of corona viruses [1, 2]; notably, the outbreak of the SARS-CoV virus in 2002 made a major health threat to the public and caused severe acute respiratory syndrome (SARS) resulted in 774 deaths over the 8098 infected cases [3, 4]. The evolution of coronavirus was continuous, in the year 2012, another version of Middle East Respiratory Syndrome-CoV (MERS-CoV) virus also emerged, and it was vulnerable; the outbreak of the MERS-CoV virus made high health emergency in the Middle East countries, infected 2506 people, and resulted in 862 deaths [5, 6]. Further, recently a new type of coronavirus also again emerged in Wuhan, China, in December 2019, which was identified as a novel coronavirus SARS-CoV-2, namely COVID-19, which causes severe acute respiratory infection (pneumonia) and other health problems [7,8,9]. As on date, nearly 4 million people were infected, in which 284,034 people died. Such high mortality rates may be due to several reasons, however, it is widely accepted that the mode of infection of SARS-CoV-2 is different from the SARS-CoV and MERS-CoV. As per reports, SARS-CoV-2 is a betacoronavirus, its genome sequence is 79% identical to SARS-CoV and 50% identical to MERS-CoV [10]. SARS-CoV-2 is an enveloped single positive-stranded RNA virus. The viral structure consists of structural proteins such as spike (S), membrane (M), envelope (E), and nucleocapsid (N). The spike S-protein present in the envelope of the virus mediates the viral particle into the host-cells. M-protein is largely present in the virion and jointly with E-protein forms a mature viral envelope. N-protein always binds with RNA, which is required for the packing of viral RNA [11, 12]. SARS-CoV-2 initiate human cell entry after the spike (S) protein present on the envelope binds to a cell membrane receptor called the angiotensin-converting enzyme (ACE2). The S-protein is cleaved into two subunits S1 and S2 by a human cell-derived protease (thought to be Furin); subsequently, S1 binds to its receptor, ACE2. The other fragment, S2, is cleaved by TMPRSS2 (transmembrane protease serine 2), a human cell surface serine protease, resulting in membrane fusion. Both ACE2 and TMPRSS2 are therefore thought to be essential in airway cells for SARS-CoV-2 infection. Inhibiting TMPRSS2 protein using suitable inhibitors can stop the virus entry and this prevents the SARS-CoV-2 virus infection. The present study is focused to find the best inhibitors of TMPRSS2 protein from the existing drugs [13,14,15,16,17] by in silico methods. Camostat mesylate, nafamostat, and leupeptin (Fig. 1) are the drugs chosen as the inhibitors of TMPRSS2, in which the drugs camostat and nafamostat are under clinical trial for COVID-19 [13, 16, 18,19,20] (https://clinicaltrials.gov/ct2/show/NCT04353284) treatment, whereas leupeptin [21] is tested for its suitability against SARS-CoV-2 viral infection and also studied for its antiviral properties against SARS-CoV viruses [22, 23]. To better understand the molecular binding mechanisms of these molecules, which are not completely known at the molecular dynamics level, we have computed the binding affinity, intermolecular interactions, and the stability of molecules from the molecular docking and the molecular dynamics simulations. The information obtained from this computational study is useful to consider these drugs as the repurposed drug for the treatment of COVID-19 disease, however it is subjected to clinical studies.
Materials and Methods
Homology Modelling and Molecular Docking
The crystal structure of TMPRSS2 protein is not available; hence, the homology model structure was generated from the amino acid sequence of TMPRSS2 with catalytic domain retrieved from the UniprotKB (Uniprot Accession O15393) using Schrödinger homology modeling interface [24]. In this homology modeling, the Hepsin structure (PDB: 5CE1) was used as a template, which was chosen based on the sequence similarity (BLAST search) [25]. The ligand molecule bound to the template structure was removed. The inbuilt secondary structure prediction (SSP) algorithm of Prime was used to predict the secondary structure of TMPRSS2. The ClustralW method incorporated to Prime was employed for aligning the sequence of TMPRSS2 with the template sequence. The unnecessary gaps found in the secondary structure were removed by manual adjustments. After the sequence alignment, the homology model of TMPRSS2 was generated. Finally, the modeled structure was optimized and refined by protein preparation wizard [26]. The structure was validated with the Ramachandran plot (Figure 1S) [27].
Preparation of Ligands and Receptor for Docking
To understand the binding affinity and the intermolecular interaction of the chosen three drug molecules camostat, nafamostat, and leupeptin [16, 18,19,20] (https://clinicaltrials.gov/ct2/show/NCT04353284) with the active site amino acids of TMPRSS2, the molecular docking simulation was performed. In prior to the docking simulation, the abovesaid three drug molecules were prepared by the Ligprep module of Schrödinger software [28]. Further, these molecules were docked with the TMPRSS2 by extra precision mode (XP) incorporated in the induced fit docking (IFD) method. Epik was employed to generate the ionization states of the ligands at pH 7.0 +/− 2.0. Thus, prepared ligand inputs were minimized using OPLS3e force field [29].
Induced Fit Docking
The induced fit docking (IFD) resulted in different conformations for each ligand-TMPRSS2 complex. Among these, the complex exhibits top docking score, lowest energy conformation, and the ligand forming expected interactions with the catalytic site residues of TMPRSS2 was selected for further studies. PyMOL software [30] was used to analyze the intermolecular interactions between the ligand and TMPRSS2 complexes.
Molecular Dynamics Simulation and Binding-Free Energy
To understand the stability, intermolecular interactions, and the binding energy of the three ligand-TMPRSS2 complexes, the molecular dynamics (MD) simulation has been performed using the OPLS3e force field [29] implemented in Desmond v5 package [31]. Further, the system was built with the pre-defined TIP4P water model and orthorhombic periodic boundary conditions at the distances 10 Å. Then, the counterions were used to neutralize the charge of these complexes with the balancing Na+/Cl− ions. Further, the constructed system for each ligand-TMPRSS2 complex was energy minimized by heating and equilibrium processes before the MD simulation. For energy minimization, the minimization and heating protocol was fixed based on the steepest descent method, annealing temperature at 0–300 K, and 2000 steps with the time steps of 0.001 ps. Further, the system was normalized in an equilibrium state at 1000 steps with a time step of 0.001 ps. Finally, the production step of the systems was continued up to 100 ns with the time steps of 0.001 ps, 300 K, and 1 atm pressure was applied using the Nose-Hoover method with NPT ensemble [32]. Intermolecular interactions and conformation of each ligand-protein complex were analyzed from the final results of MD simulation. Among the 1000 fractions, the 10, 20, 30, 40, 50, 60, 70, 80, 90, and 100 ns fractions were used to determine the binding-free energy (MM/GBSA) of the ligand-TMPRSS2 complexes by Prime application incorporated to Schrödinger software package [33].
Results and Discussion
Molecular Docking Simulation and Ligand(s)-TMPRSS2 Binding
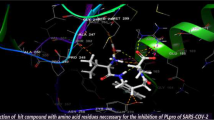
TMPRSS2 is the serine protease, which mediates the S-protein of SARS-CoV-2 for the virus entry through the ACE2 enzyme. Reportedly, by inhibiting the TMPRSS2 protease, the SARS-CoV-2 fusion can be stopped [13,14,15,16,17]. In TMPRSS2, the ligand-binding active site has catalytic triad consists of Ser186, His41, and Asp180 amino acids; to inhibit the function of this enzyme, ligands necessarily bind with these key amino acids. To understand the binding mechanism, initially, the molecular docking simulations for the drug molecules camostat, nafamostat, and leupeptin with TMPRSS2 have been performed using induced fit IFD method. The docking score values of the three molecules are − 6.648, − 7.075, and − 9.325 kcal/mol, respectively, whereas the IFD scores are − 490.01, − 493.20, and − 501.23 kcal/mol; and the glide energy values are − 57.457, − 56.96, and − 64.35 kcal/mol (Table 1). On comparing the abovesaid values reveal that the leupeptin consistently exhibits high score. The magnitude of difference in docking scores and glide energy reflects the nature of their intermolecular interactions with the neighboring amino acids present in the active site of TMPRSS2 protease (Fig. 2). In the catalytic site, the camostat and leupeptin are forming strong interactions with Asp180, Ser186, and His41 than the nafamostat (Table 2); however, the interactions of leupeptin with the catalytic amino acids Asp180, Ser186, and His41are found stronger and the interaction distances are 1.9, 2.1, 2.0, and 3.5 Å, respectively. Figure 3 shows the superimposed form of the three drug molecules in the active site of TMPRSS2 revealing the orientation of the molecules and differences. From this, we observed that these three molecules have a tendency to bind with the serine 2 protease TMPRSS2. However, the nafamostat is lacking the Ser186 and His41 interactions with TMPRSS2 (Table 2). The intermolecular interactions of camostat, nafamostat, and leupeptin with the catalytic triad of TMPRSS2 were compared with the reported complexes [34, 35] [Table 3]. In the present study, the camostat molecule forms intermolecular interactions with the expected three catalytic triad residues (His41, Asp180, Ser186), whereas the reported complex [34] exhibits only two interactions with the active site catalytic triad residues (His296 and Ser441) of TMPRSS2. And the nafamostat molecule forms only one interaction with the catalytic triad residue (Asp180), which is unlike the reported complex [34] (His296). Interestingly, leupeptin molecule forms interaction with all the residues (His41, Asp180, Ser186) of catalytic triad, whereas the reported structure [35] shows leupeptin forms two interactions with the catalytic triad residues (His57 and Ser214). From this comparison, it is clear that how the three molecules of the present study forms interactions with the active site catalytic triad of TMPRSS2 and also how these interactions differ from the reported structures. Further, this study also probed the sustainability of these interactions and the binding affinity of the three drug molecules with the TMPRSS2 during the molecular dynamics simulations.
Stability of Ligand(s)-TMPRSS2 Complexes and Intermolecular Interactions
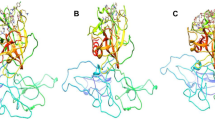
Molecular dynamics simulation is the computational technique, which allows to understand the biological function of proteins. Here, we performed 100 ns MD simulation for the three ligand-TMPRSS2 complexes; the results explore the stability and the binding affinity of the three drug molecules towards the TMPRSS2. Figure 4 and 5 displays the variation of RMSD and RMSF values of the three complexes during the MD simulation. The RMSD values of the molecules camostat, nafamostat, and leupeptin remain within 2 Å (Fig. 4(a-c), whereas the RMSD of protein in the camostat-TMPRSS2 and nafamostat-TMPRSS2 complexes are found high comparing with the leupeptin bound TMPRSS2 complex (2.4 Å). Figure 5(a-c) displays the variations in the RMSF of TMPRSS2 of three complexes; among these, a strong fluctuation is found in the region of residue number 60 of camostat-TMPRSS2 complex and no such incidence is noticed in the RMSF map of other two complexes. The superimposed form of the three ligand-TMPRSS2 complexes (Fig. 6) obtained from the docking and MD simulations is showing the conformational difference of ligands and TMPRSS2 protein in the respective complex; specifically, this allows to visualize that how the conformation and the orientation of the ligands and proteins are altered during the MD simulation. Overall, the fluctuation of the leupeptin molecule is found to be very less and it is normal. Furthermore, surprisingly, the superimposed form of docked and MD complexes of leupeptin (Fig. 6) are showing not much deviation, whereas the other two complexes deviated considerably, this confirms that leupeptin is very stable while it binds with the TMPRSS2 protein.
The intermolecular interactions between the ligand and the active site amino acids of TMPRSS2 of three complexes obtained from the MD simulations are shown in Fig. 7(a-c). On comparing these interactions with the interactions of ligand-TMPRSS2 complexes (Fig. 2) from docking simulation reveals that during the MD simulation, some interactions became strong and in addition that few new interactions also formed (Fig. 7). Further, when we examine the interactions present in the camostat-TMPRSS2 complex, the terminal NH2 group of camostat molecule forms interactions with one of the key amino acids Asp180 at the distances 1.6 and 1.7 Å, and these interactions are also very stable during the MD simulation, whereas in the nafamostat-TMPRSS2 complex, we observed a similar trend, in which the molecule forms interactions with Asp180 at the distances 2.0 and 1.9 Å; and it also forms an interaction with His41, the contact distance is 2.3 Å. Furthermore, these two molecules also form several interactions with the nearby amino acids present in the active site. In the leupeptin-TMPRSS2 complex, as found in the docked complex, the MD simulation also displays the same hydrogen bonding interactions between leupeptin and the key amino acids Asp180, His41, and Ser186 at the distances, 1.8, 1.8, and 1.7 Å. These important interactions were highly stable throughout the 100-ns MD simulations and this can be visualized from Figs. 8(a-c), 9(a-c) and Figure 2S. The characterization of interactions between the drug molecules and TMPRSS2 reveals that the leupeptin molecule is highly stable and forms the expected interactions with the key amino acids Asp180, His41, and Ser186 of catalytic site and found very strong, whereas the other two drug molecules, camostat and nafamostat, are lacking the interactions with His41 and Ser186 amino acids. The binding energy for the three complexes was calculated using MM-GBSA method. The calculated values of the binding energy of camostat, nafamostat, and leupeptin molecules with TMPRESS2 are − 42.77, − 52.86, and − 55.25 kcal/mol. Among these molecules, relatively, leupeptin exhibits high binding affinity with the TMPRSS2 serine protease. Overall, leupeptin has high stability and forms very strong interactions with the key amino acids compare with the other two drug molecules.
Conclusions
To address the urgent need of drugs to treat the COVID-19 pandemic disease, repurposing drugs is one of the best solutions comparing with the other techniques like vaccine development and new plasma transfusion. In the line of drug searching process, we have undertaken camostat, nafamostat, and leupeptin drug molecules to inhibit the TMPRSS2 serine 2 protease, which is the activator of SARS-CoV-2 for the fusion of the virus enter into the host human cell. The molecular docking study reveals that these three drug molecules form interactions and has binding affinity towards TMPRSS2. However, the leupeptin forms strong interactions with the key amino acids Ser186, His41, and Asp180 of catalytic triad present in the active site of TMPRSS2 as the other two molecules lacking Ser186 and His41 interactions, whereas in the molecular dynamics simulations of these three complexes, leupeptin is highly stable (RMSD & RMSF) and this molecule also forms stable interactions with the key amino acids Ser186, His41, and Asp186 of TMPRSS2 during the 100-ns MD simulations. On comparing the binding-free energy of all the three molecules, notably, leupeptin has high binding affinity towards TMPRSS2. From the above static and dynamic studies, it is confirmed that leupeptin is very stable and it could strongly inhibit the TRMPRSS2 serine 2 protease and this leads to stop the fusion of SARS-CoV-2 virus onto the host human cell, hence it is considered as a repurposed drug after clinical studies.
Data Availability
Not applicable
References
Woo, P. C., Huang, Y., Lau, S. K., & Yuen, K. Y. (2010). Coronavirus genomics and bioinformatics analysis. Viruses, 2(8), 1804–1820.
Woo, P. C., Lau, S. K., Lam, C. S., Lau, C. C., Tsang, A. K., Lau, J. H., Bai, R., Teng, J. L., Tsang, C. C., Wang, M., Zheng, B. J., Chan, K. H., & Yuen, K. Y. (2012). Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. Journal of Virology, 86(7), 3995–4008.
Kuiken, T., Fouchier, R. A., Schutten, M., Rimmelzwaan, G. F., van Amerongen, G., van Riel, D., Laman, J. D., de Jong, T., van Doornum, G., Lim, W., Ling, A. E., Chan, P. K., Tam, J. S., Zambon, M. C., Gopal, R., Drosten, C., van der Werf, S., Escriou, N., Manuguerra, J. C., Stöhr, K., Peiris, J. S., & Osterhaus, A. D. (2003). Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. The Lancet, 362(9380), 263–270.
Zhong, N. S., Zheng, B. J., Li, Y. M., Poon, Xie, Z. H., Chan, K. H., Li, P. H., Tan, S. Y., Chang, Q., Xie, J. P., Liu, X. Q., Xu, J., Li, D. X., Yuen, K. Y., Peiris, & Guan, Y. (2003). Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. The Lancet, 362(9393), 1353–1358.
Zumla, A., Hui, D. S., & Perlman, S. (2015). Middle East respiratory syndrome. The Lancet, 386(9997), 995–1007.
Aleanizy, F. S., Mohmed, N., Alqahtani, F. Y., & El Hadi Mohamed, R. A. (2017). Outbreak of Middle East respiratory syndrome coronavirus in Saudi Arabia: a retrospective study. BMC Infectious Diseases, 17(1), 23.
Wang, C., Horby, P. W., Hayden, F. G., & Gao, G. F. (2020). A novel coronavirus outbreak of global health concern. The Lancet, 395(10223), 470–473.
Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J., Hu, Y., Zhang, L., Fan, G., Xu, J., Gu, X., Cheng, Z., Yu, T., Xia, J., Wei, Y., Wu, W., Xie, X., Yin, W., Li, H., Liu, M., Xiao, Y., Gao, H., Guo, L., Xie, J., Wang, G., Jiang, R., Gao, Z., Jin, Q., Wang, J., & Cao, B. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet, 395(10223), 497–506.
Zhou, P., Yang, X. L., Wang, X. G., Hu, B., Zhang, L., Zhang, W., Si, H. R., Zhu, Y., Li, B., Huang, C. L., Chen, H. D., Chen, J., Luo, Y., Guo, H., Jiang, R. D., Liu, M. Q., Chen, Y., Shen, X. R., Wang, X., Zheng, X. S., Zhao, K., Chen, Q. J., Deng, F., Liu, L. L., Yan, B., Zhan, F. X., Wang, Y. Y., Xiao, G. F., & Shi, Z. L. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579(7798), 270–273.
Lu, R., Zhao, X., Li, J., Niu, P., Yang, B., Wu, H., Wang, W., Song, H., Huang, B., Zhu, N., Bi, Y., Ma, X., Zhan, F., Wang, L., Hu, T., Zhou, H., Hu, Z., Zhou, W., Zhao, L., Chen, J., Meng, Y., Wang, J., Lin, Y., Yuan, J., Xie, Z., Ma, J., Liu, W. J., Wang, D., Xu, W., Holmes, E. C., Gao, G. F., Wu, G., Chen, W., Shi, W., & Tan, W. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. The Lancet, 395(10224), 565–574.
Neuman, B. W., Adair, B. D., Yoshioka, C., Quispe, J. D., Orca, G., Kuhn, P., Milligan, R. A., Yeager, M., & Buchmeier, M. J. (2006). Supramolecular architecture of severe acute respiratory syndrome coronavirus revealed by electron cryomicroscopy. Journal of Virology, 80(16), 7918–7928.
Bárcena, M., Oostergetel, G. T., Bartelink, W., Faas, F. G., Verkleij, A., Rottier, P. J., Koster, A. J., & Bosch, B. J. (2009). Cryo-electron tomography of mouse hepatitis virus: insights into the structure of the coronavirion. PNAS, 106(2), 582–587.
Hoffmann, M., Kleine-Weber, H., Krüger, N., Müller, M., Drosten, C., & Pöhlmann, S. (2020). The novel coronavirus 2019 (COVID-19) uses the SARS-1 coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. BioRxiv. Preprint. https://doi.org/10.1101/2020.01.31.929042.
Glowacka, I., Bertram, S., Muller, M. A., Allen, P., Soilleux, E., Pfefferle, S., Steffen, I., Tsegaye, T. S., He, Y., Gnirss, K., Niemeyer, D., Schneider, H., Drosten, C., & Pöhlmann, S. (2011). Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus Spike protein for membrane fusion and reduces viral control by the humoral immune response. Journal of Virology, 85(9), 4122–4134.
Iwata-Yoshikawa, N., Okamura, T., Shimizu, Y., Hasegawa, H., Takeda, M., & Nagata, N. (2019). TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. Journal of Virology, 93(6), e01815-18.
Kawase, M., Shirato, K., van der Hoek, L., Taguchi, F., & Matsuyama, S. (2012). Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitorsprevents severe acute respiratory syndrome coronavirus entry. Journal of Virology, 86(12), 6537–6654.
Zhou, Y., Vedantham, P., Lu, K., Agudelo, J., Carrion Jr., R., Nunneley, J. W., Barnard, D., Pöhlmann, S., McKerrow, J. H., Renslo, A. R., & Simmons, G. (2015). Protease inhibitors targeting coronavirus and filovirus entry. Antiviral Research, 116, 76–84.
Bittmann, S., Luchter, E., Weissenstein, A., Villalon, G., & Moschüring-Alieva, E. (2020). TMPRSS2-inhibitors play a role in cell entry mechanism of COVID-19: an insight into camostat and nafamostat. Journal of Regenerative Biology and Medicine, 2(2), 1–3.
Hoffmann, M., Schroeder, S., Kleine-Weber, H., Müller, M. A., Drosten, C., & Pöhlmann, S. (2020). Nafamostat mesylate blocks activation of SARS-CoV-2: new treatment option for COVID-19. Antimicrobial Agents and Chemotherapy. (In press). https://doi.org/10.1128/AAC.00754-20.
Sonawane, K. D., Barale, S. S., Dhanavade, M. J., Waghmare, S. R., Nadaf, N. H., Kamble, S. A., Mohammed, A. A., Makandar, A. M., Fandilolu, P. M., Dound, A. S., & Naik, N. M. (2020). Homology modeling and docking studies of TMPRSS2 with experimentally known inhibitors camostatmesylate, nafamostat and bromhexinehydrochloride to control SARS-Coronavirus-2. ChemRxiv. Preprint. https://doi.org/10.26434/chemrxiv.12162360.v1.
Appleyard, G., & Tisdale, M. (1985). Inhibition of the growth of human coronavirus 229E by leupeptin. Journal of General Virology, 66(2), 363–366.
Mittal, L., Kumari, A., Srivastava, M., Singh, M., & Asthana, S. (2020). Identification of potential molecules against COVID-19 main protease through structure-guided virtual screening approach. ChemRxiv. Preprint. https://doi.org/10.26434/chemrxiv.12086565.v2.
Simmons, G., Gosalia, D., Rennekamp, A., Reeves, J., Diamond, S., & Bates, P. (2005). Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proceedings of the National Academy of Sciences of the United States of America, 102, 11876–11881. https://doi.org/10.1073/pnas.0505577102.
(2020). Schrödinger Release 2020-1: Prime, Schrödinger, LLC, New York, NY.
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., & Lipman, D. J. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215(3), 403–410.
(2012). Schrödinger Suite 2012 Protein Preparation Wizard; Epik version 2.3, Schrödinger, LLC, New York, NY, 2012; Impact version 5.8, Schrödinger, LLC, New York, NY, 2012; Prime version 3.1, Schrödinger, LLC, New York, NY.
Ramachandran, G. N., Ramakrishnan, C., & Sasisekharan, V. (1963). Stereochemistry of polypeptide chain configurations. Journal of Molecular Biology, 7, 95–99.
(2020). Schrödinger Release 2020-1: LigPrep, Schrödinger, LLC, New York, NY.
Harder, E., Damm, W., Maple, J., Wu, C., Reboul, M., Xiang, J. Y., Wang, L., Lupyan, D., Dahlgren, M. K., Knight, J. L., Kaus, J. W., Cerutti, D., Krilov, G., Jorgensen, W. L., Abel, R., & Friesner, R. A. (2016). OPLS3: a force field providing broad coverage of drug-like small molecules and proteins. Journal of Chemical Theory Computation, 12(1), 281–296.
The PyMOL (2002). Molecular Graphics System. Version 2.0 Schrödinger, LLC.
Desmond Molecular Dynamics System, D. E. Shaw Research, New York, NY. (2020). Maestro-Desmond Interoperability Tools. New York: Schrödinger.
Evans, D. J., & Holian, B. L. (1985). The Nose–Hoover thermostat. The Journal of Chemical Physics, 83(8), 4069–4074.
Li, J., Abel, R., Zhu, K., Cao, Y., Zhao, S., & Friesner, R. A. (2011). The VSGB 2.0 model: a next generation energy model for high resolution protein structure modelling. Proteins, 79, 4–812.
Mukhtar, O. I., Abeeb, A. Y., Oluwaseun, S. A., & Olanrewaju, A. D. (2020). Computer-aided screening for potential TMPRSS2 inhibitors: a combination of pharmacophore modeling, molecular docking and molecular dynamics simulation approaches. Journal of Biomolecular Structure and Dynamics. https://doi.org/10.1080/07391102.2020.1792346.
Kurinov, I. V., & Harrison, R. W. (1996). Two crystal structures of the leupeptin-trypsin complex. Protein Science, 5(4), 752–758.
Acknowledgements
The authors thank the Computer Centre, Periyar University, Salem, for performing the computational work in the High Performance Cluster (HPC) Computer.
Author information
Authors and Affiliations
Contributions
Jaganathan Ramakrishnan, Saravanan Kandasamy, Kumaradhas Poomani designed the research; Jaganathan Ramakrishnan, Saravanan Kandasamy, Ancy Iruthayaraj, Sivanandam Magudeeswaran, Kalaiarasi Chinnasamy, Kumaradhas Poomani performed the Homology modeling, molecular docking, and molecular dynamics simulation, and analyzed the results. The authors are equally contributed to prepare and reviewed the manuscript.
Corresponding author
Ethics declarations
Ethical Approval
Not applicable
Consent to Participate
Not applicable
Consent to Publish
All authors consented to the publication of this work. Authors all confirm the permission of publication for this study.
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(DOC 480 kb)
Rights and permissions
About this article
Cite this article
Ramakrishnan, J., Kandasamy, S., Iruthayaraj, A. et al. Strong Binding of Leupeptin with TMPRSS2 Protease May Be an Alternative to Camostat and Nafamostat for SARS-CoV-2 Repurposed Drug: Evaluation from Molecular Docking and Molecular Dynamics Simulations. Appl Biochem Biotechnol 193, 1909–1923 (2021). https://doi.org/10.1007/s12010-020-03475-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-020-03475-8