Abstract
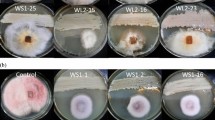
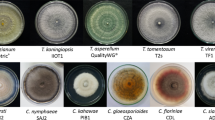
In this study, medicinal plant (Solanum surattense)-associated bacteria were isolated and their extracellular secondary metabolites were extracted. Dual-plate application of crude secondary metabolites proved that SSL2I and SSL5 had a good inhibitory activity against Ralstonia solanacearum. These biocontrol bacteria were identified as Bacillus subtilis and Bacillus velezensis by 16S rRNA gene sequencing analysis. The crude extracts of secondary metabolites were identified based on high-resolution liquid chromatography/mass spectrometry (HR-LCMS) analysis. On the basis of HR-LCMS analysis, we selected the compounds such as glucosamine and indole acetaldehyde for in silico analysis and inhibition of pathogenic gene of phcA from R. solanacearum. The specificity of identified pathogenic gene of R. solanacearum and its cytoplasmic localization were identified by BLASTP and PSORTB bioinformatics tools. The protein–protein interaction between the identified secondary metabolites and pathogenic gene revealed that the compound had antagonistic potential against pathogenic gene of phcA. Furthermore, the synthetic forms of the above metabolites showed that both the compounds had the ability to inhibit R. solanacearum under in vitro condition. On the basis of in silico and in vitro analyses, it was concluded that medicinal plant-associated Bacillus spp. could be used as a biocontrol agent in managing wilt disease caused by R. solanacearum.




Similar content being viewed by others
References
Poueymiro, M., Cazale, A. C., Francois, J. M., Parrou, J. L., Peeters, N., & Genin, A. (2014). Ralstonia solanacearum type III effector directs the production of the plant signal metabolite trehalose-6-phosphate. MBio, 5, e02065–e02014.
Coutinho, T. A., & Wingfield, M. J. (2017). Ralstonia solanacearum and R. pseudosolanacearum on eucalyptus: opportunists or primary pathogens? Frontiers in Plant Science, 8, 761.
Yuliar, Nion, Y. A., & Toyota, K. (2015). Recent trends in control methods for bacterial wilt diseases caused by Ralstonia solanacearum. Microbes and Environments, 30(1), 1–11.
Meng, F., Babujee, L., Jacobs, J. M., & Allen, C. (2015). Comparative transcriptome analysis reveals cool virulence factors of Ralstonia solanacearum race 3 biovar 2. PLoS One, 10(10), e0139090.
Yang, L., Li, S., Qin, X., Jiang, G., Chen, J., Li, B., Yao, X., Liang, P., Zhang, Y., & Ding, W. (2017). Exposure to umbelliferone reduces Ralstonia solanacearum biofilm formation, transcription of type III secretion system regulators and effectors and virulence on tobacco. Frontiers in Microbiology, 8, 1234.
Khokhani, D., Lowe-Power, T. M., Tran, T. M., & Allen, C. (2017). A single regulator mediates strategic switching between attachment/spread and growth/virulence in the plant pathogen Ralstonia solanacearum. MBio, 8, e00895–e00817.
Huang, J., Yindeeyoungyeon, W., Garg, R. P., Denny, T. P., & Schell, M. A. (1998). Joint transcriptional control of XpsR, the unusual signal integrator of the Ralstonia solanacearum virulence gene regulatory network, by a response regulator and a LysR-type transcriptional activator. Journal of Bacteriology, 180(10), 2736–2743.
Ravindra, M., & Vijay Kumar, S. (2008). Evaluation of antifungal property of medicinal plants. Journal of Phytological Research, 21, 139–142.
Li, H., Guan, Y., Dong, Y., Zhao, L., Rong, S., Chen, W., Lv, M., Xu, H., Gao, X., Chen, R., & Li, L. (2018). Isolation and evaluation of endophytic Bacillus tequilensis GYLH001 with potential application for biological control of Magnaporthe oryzae. PLoS One, 13(10), e0203505.
Mohamad, A., Abdalla, O., Li, L., Ma, J., Hatab, S. R., Xu, L., Guo, J. W., Rasulov, B. A., Liu, Y. H., Hedlund, B. P., & Li, W. J. (2018). Evaluation of the antimicrobial activity of endophytic bacterial populations from Chinese traditional medicinal plant licorice and characterization of the bioactive secondary metabolites produced by Bacillus atrophaeus against Verticillium dahlia. Frontiers in Microbiology, 9, 924.
Kaki, A. A., Chaouche, N. K., Dehimat, L., Milet, A., Youcef-Ali, M., Ongena, M., & Thonart, P. (2013). Biocontrol and plant growth promotion characterization of Bacillus species isolated from Calendula officinalis rhizosphere. Indian Journal of Microbiology, 53(4), 447–452.
Ausubel, F. M., Brent, R., Robert, E., Kingston Moore, D. D., Seidman, J. G., Smith, J. A., & Struhl, K. (1992). Current protocols in molecular biology. New York: Greene Publishing Association Wiley-Interscience.
Yasmin, S., Hafeez, F. Y., Mirza, M. S., Rasul, M., Arshad, H. M., Zubair, M., & Iqbal, M. (2017). Biocontrol of bacterial leaf blight of rice and profiling of secondary metabolites produced by rhizospheric Pseudomonas aeruginosa BRp3. Frontiers in Microbiology, 8, 1895.
Priyanka, S. R., Jinal, H. N., & Amaresan, N. (2018). Diversity and antimicrobial activity of plant associated bacteria from selected medicinal plants in Kutch, Dhinodhar hill, Gujarat. National Academy Science Letters, 41(3), 137–139.
Jinal, H. N., & Amaresan, N. (2019). Characterization of medicinal plant-associated biocontrol Bacillus subtilis (SSL2) by liquid chromatography-mass spectrometry and evaluation of compounds by in silico and in vitro methods. Journal of Biomolecular Structure and Dynamics, 38(2), 500–510. https://doi.org/10.1080/07391102.2019.1581091.
Yu, N. Y., Wagner, J. R., Laird, M. R., Melli, G., Rey, S., Lo, R., Dao, P., Sahinalp, S. C., Ester, M., Foster, L. J., & Brinkman, F. S. (2010). PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics, 26(13), 1608–1615.
Munikumar, M., Priyadarshini, I. V., Pradhan, D., Sandeep, S., Umamaheswari, A., & Vengamma, B. (2012). In silico identification of common putative drug targets among the pathogens of bacterial meningitis. Biochemistry and Analytical Biochemistry, 1, 123.
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., & Lipman, D. J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research, 25(17), 3389–3402.
Reddy, K. K., Singh, S. K., Tripathi, S. K., Selvaraj, C., & Suryanarayanan, V. (2013). Shape and pharmacophore-based virtual screening to identify potential cytochrome P450 sterol 14α-demethylase inhibitors. Journal of Receptors and Signal Transduction, 33(4), 234–243.
Selvaraj, C., Krishnasamy, G., Jagtap, S. S., Patel, S. K., Dhiman, S. S., Kim, T. S., Singh, S. K., & Lee, J. K. (2016). Structural insights into the binding mode of d-sorbitol with sorbitol dehydrogenase using QM-polarized ligand docking and molecular dynamics simulations. Biochemical Engineering Journal, 114, 244–256.
Singh, G., & Singh, V. (2018). Functional elucidation of hypothetical proteins for their indispensable roles toward drug designing targets from Helicobacter pylori strain HPAG1. Journal of Biomolecular Structure Dynamics, 36(4), 906–918.
Selvaraj, C., Sivakamavalli, J., Vaseeharan, B., Singh, P., & Singh, S. K. (2014). Examine the characterization of biofilm formation and inhibition by targeting SrtA mechanism in Bacillus subtilis: a combined experimental and theoretical study. Journal of Molecular Modeling, 20(8), 2364–2368.
Karlsson, E., Shin, J. H., Westman, G., Eriksson, L. A., Olsson, L., & Mapelli, V. (2018). In silico and in vitro studies of the reduction of unsaturated α,β bonds of trans-2-hexenedioic acid and 6-amino-trans-2-hexenoic acid – Important steps towards biobased production of adipic acid. PLoS One, 13(2), e0193503.
Qin, C., Tao, J., Liu, T., Liu, Y., Xiao, N., Li, T., Gu, Y., Yin, H., & Meng, D. (2019). Responses of phyllosphere microbiota and plant health to application of two different biocontrol agents. AMB Express, 9(1), 42.
Zhao, Z., Wang, Q., Wang, K., Brian, K., Liu, C., & Gu, Y. (2010). Study of the antifungal activity of Bacillus vallismortis ZZ185 in vitro and identification of its antifungal components. Bioresource Technology, 101(1), 292–297.
Dimkić, Zivkovic, S., Beric, T., Ivanovic, Z., Gavrilovic, V., Stankovic, S., & Fira, D. (2013). Characterization and evaluation of two Bacillus strains, SS-12.6 and SS-13.1, as potential agents for the control of phytopathogenic bacteria and fungi. Biological Control, 65(3), 312–321.
Tiwari, S., Jamal, S. B., Hassan, S. S., Carvalho, P. V., Almeida, S., Barh, D., Ghosh, P., Silva, A., Castro, T. L., & Azevedo, V. (2017). Two-component signal transduction systems of pathogenic bacteria as targets for antimicrobial therapy: an overview. Frontiers in Microbiology, 8, 878.
Schaefers, M. M., Liao, T. L., Boisvert, N. M., Roux, D., Yoder-Himes, D., & Priebe, G. P. (2017). An oxygen-sensing two-component system in the Burkholderia cepacia complex regulates biofilm, intracellular invasion, and pathogenicity. PLoS Pathogens, 13(1), e1006116.
Genin, S., & Denny, T. P. (2012). Pathogenomics of the Ralstonia solanacearum species complex. Annual Review of Phytopathology, 50(1), 67–89.
Genin, S., & Boucher, C. (2002). Ralstonia solanacearum: secrets of a major pathogen unveiled by analysis of its genome. Molecular Plant Pathology, 3(3), 111–118.
Liu, H., Zhang, S., Schell, M. A., & Denny, T. P. (2005). Pyramiding unmarked deletions in Ralstonia solanacearum shows that secreted proteins in addition to plant cell-wall-degrading enzymes contribute to virulence. Molecular Plant-Microbe Interaction, 18(12), 1296–1305.
Acharya, A., & Garg, L. C. (2016). Drug target identification and prioritization for treatment of ovine foot rot: an in silico approach. International Journal of Genomics. https://doi.org/10.1155/2016/7361361.
Patil, R., Das, S., Stanley, A., Yadav, L., Sudhakar, A., & Varma, A. K. (2010). Optimized hydrophobic interactions and hydrogen bonding at the target-ligand interface leads the pathways of drug-designing. PLoS One, 5(8), e12029.
Acknowledgments
The authors thank the management of UTU and Director, CGBIBT for support and providing facility to carry out the work. The authors are also grateful to IIT Bombay—SAIF for high-resolution LC-MS analysis studies.
Funding
This study received financial support from the B. U. Patel Doctoral Fellowship Scheme, Uka Tarsadia University (UTU), Bardoli, Gujarat, India.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Jinal, H.N., Amaresan, N. In Silico and In Vitro Analyses of Glucosamine and Indole Acetaldehyde Inhibit Pathogenic Regulator Gene phcA of Ralstonia solanacearum, a Causative Agent of Bacterial Wilt of Tomato. Appl Biochem Biotechnol 192, 230–242 (2020). https://doi.org/10.1007/s12010-020-03328-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-020-03328-4