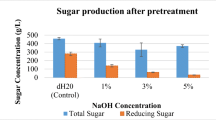
Abstract
We evaluated various agricultural lignocellulosic biomass and variety of fungi to produce cellulolytic enzymes cocktail to yield high amount of reducing sugars. Solid-state fermentation was performed using water hyacinth, paddy straw, corn straw, soybean husk/tops, wheat straw, and sugarcane bagasse using fungi like Nocardiopsis sp. KNU, Trichoderma reesei, Trichoderma viride, Aspergillus flavus, and Phanerochaete chrysosporium alone and in combination to produce cellulolytic enzymes. Water hyacinth produced (U ml−1) endoglucanase (51.13) and filter paperase (0.55), and corn straw produced (U ml−1) β-glucosidase (4.65), xylanase (113.32), and glucoamylase (41.27) after 7-day incubation using Nocardiopsis sp. KNU. Production of cellulolytic enzymes was altered due to addition of various nitrogen sources, metal ions, vitamins, and amino acids. The maximum cellulolytic enzymes were produced by P. chrysosporium (endoglucanase; 166.32 U ml−1 and exoglucanase; 12.20 U ml−1), and by T. viride (filter paperase; 1.57 U ml−1). Among all, co-culture of T. reesei, T. viride, A. flavus, and P. chrysosporium showed highest β-glucosidase (17.05 U ml−1). The highest xylanase (1129 U ml−1) was observed in T. viride + P. chrysosporium co-culture. This study revealed the dependency on substrate and microorganism to produce good quality enzyme cocktail to obtain maximum reducing sugars.




Similar content being viewed by others
References
Armaroli, N., & Balzani, V. (2011). Towards an electricity-powered world. Energy and Environmental Science, 4, 3193–3222.
Dlangamandla, N., Ntwampe, S. K. O., Angadam, J. O., Itoba-Tombo, E. F., Chidi, B. S., & Mekuto, L. (2019). Integrated hydrolysis of mixed agro-waste for a second generation biorefinery using Nepenthes mirabilis pod digestive fluids. Processes, 7, 64.
Biswas, R., Zheng, T., Olson, D. G., Lynd, L. R., & Guss, A. M. (2015). Elimination of hydrogenase active site assembly blocks H2 production and increases ethanol yield in Clostridium thermocellum. Biotechnology for Biofuels, 8, 20.
Kralova, I., & Sjoblom, J. (2010). Biofuels-renewable energy sources: A review. Journal of Dispersive Science and Technology, 31, 409–425.
Sun, Y., & Cheng, J. Y. (2002). Hydrolysis of lignocellulosic materials for ethanol production: A review. Bioresource Technology, 83(1), 1–11.
Gandla, M. L., Martín, C., & Jönsson, L. J. (2018). Analytical enzymatic saccharification of lignocellulosic biomass for conversion to biofuels and bio-based chemicals. Energies, 11, 2936.
Dhillon, G. S., Kaur, S., Brar, S. K., & Verma, M. (2012). Potential of apple pomace as a solid substrate for fungal cellulase and hemicellulase bioproduction through solid-state fermentation. Industrial Crops and Products, 38, 6–13.
Jung, S., Song, Y., Kim, H. M., & Bae, H. J. (2015). Enhanced lignocellulosic biomass hydrolysis by oxidative lytic polysaccharide monooxygenases (LPMOs) GH61 from Gloeophyllum trabeum. Enzyme and Microbial Technology, 77, 38–45.
Kumar, B., Trivedi, P., Mishra, A. K., Pandey, A., & Palni, L. M. S. (2004). Microbial diversity of soil from two hot springs in Uttaranchal Himalaya. Microbiological Research, 159(2), 141–146.
Khan, M. M. H., Ali, S., Fakhru'L-Razi, A., & Alam, M. Z. (2007). Use of fungi for the bioconversion of rice straw into cellulase enzyme. Journal of Environmental Science and Health Part B-Pesticides Food Contamination and Agricultural Wastes, 42, 381–386.
Bansal, N., Tewari, R., Soni, R., & Soni, S. K. (2012). Production of cellulases from Aspergillus niger NS-2 in solid state fermentation on agricultural and kitchen waste residues. Waste Management, 32, 1341–1346.
Maeda, R. N., Barcelos, C. A., Anna, L. M. M. S., & Pereira, N. (2013). Cellulase production by Penicillium funiculosum and its application in the hydrolysis of sugar cane bagasse for second generation ethanol production by fed batch operation. Journal of Biotechnology, 163(1), 38–44.
Salihu, A., Abbas, O., Sallau, A. B., & Alam, M. Z. (2015). Agricultural residues for cellulolytic enzyme production by Aspergillus niger: Effects of pretreatment. 3 Biotech, 5, 1101–1106.
Ferraz, J. L. A. A., Souza, L. O., Soares, G. A., Coutinho, J. P., de Oliveira, J. R., Oliveira, E. A., & Franco, M. (2018). Enzymatic saccharification of lignocellulosic residues using cellulolytic enzyme extract produced by Penicillium roqueforti ATCC 10110 cultivated on residue of yellow mombin fruit. Bioresource Technology, 248, 214–220.
Santos, T. C., Reis, N. S., Silva, T. P., Machado, F. P. P., Bonomo, R. C. F., & Franco, M. (2016). Prickly palm cactus husk as a raw material for production of ligninolytic enzymes by Aspergillus niger. Food Science and Biotechnology, 25(1), 205–211.
Marques, G. L., dos Santos Reis, N., Silva, T. P., Ferreira, M. L. O., Oliveira, E. A., Oliveira, J. R., & Franco, M. (2018). Production and characterisation of xylanase and endoglucanases produced by Penicillium roqueforti ATCC 10110 through the solid-state fermentation of rice husk residue. Waste and Biomass Valorization, 9, 2061–2069.
Singhania, R. R., Saini, J. K., Saini, R., Adsul, M., Mathur, A., Gupta, R., & Tuli, D. K. (2014). Bioethanol production from wheat straw via enzymatic route employing Penicillium janthinellum cellulases. Bioresource Technology, 169, 490–495.
Yoon, L. W., Ang, T. N., Ngoh, G. C., & Chua, A. S. M. (2014). Fungal solid-state fermentation and various methods of enhancement in cellulase production. Biomass and Bioenergy, 67, 319–338.
Tanaka, H., Koike, K., Itakura, S., & Enoki, A. (2009). Degradation of wood and enzyme production by Ceriporiopsis subvermispora. Enzyme and Microbial Technology, 45, 384–390.
Shrestha, P., Khanal, S. K., Pometto, A. L., & van Leeuwen, J. (2009). Enzyme production by wood-rot and soft-rot fungi cultivated on corn fiber followed by simultaneous saccharification and fermentation. Journal of Agricultural and Food Chemistry, 57(10), 4156–4161.
Lo, C. M., Zhang, Q., Callow, N. V., & Ju, L. K. (2010). Cellulase production by continuous culture of Trichoderma reesei Rut C30 using acid hydrolysate prepared to retain more oligosaccharides for induction. Bioresource Technology, 101(2), 717–723.
Singhania, R. R., Sukumaran, R. K., Patel, A. K., Larroche, C., & Pandey, A. (2010). Advancement and comparative profiles in the production technologies using solid-state and submerged fermentation for microbial cellulases. Enzyme and Microbial Technology, 46, 541–549.
Saratale, G. D., Saratale, R. G., & Oh, S. E. (2012). Production and characterization of multiple cellulolytic enzymes by isolated Streptomyces sp. MDS. Biomass and Bioenergy, 47, 302–315.
Qi, B. K., Chen, X. R., Shen, F., Su, Y., & Wan, Y. H. (2009). Optimization of enzymatic hydrolysis of wheat straw pretreated by alkaline peroxide using response surface methodology. Industrial and Engineering Chemistry Research, 48, 7346–7353.
Jeya, M., Zhang, Y. W., Kim, I. W., & Lee, J. K. (2009). Enhanced saccharification of alkali-treated rice straw by cellulase from Trametes hirsuta and statistical optimization of hydrolysis conditions by RSM. Bioresource Technology, 100(21), 5155–5161.
Kshirsagar, S. D., Waghmare, P. R., Loni, P. C., Patil, S. A., & Govindwar, S. P. (2015). Dilute acid pretreatment of rice straw, structural characterization and optimization of enzymatic hydrolysis conditions by response surface methodology. RSC Advances, 5, 46525–46533.
Chen, M., Zhao, J., & Xia, L. M. (2008). Enzymatic hydrolysis of maize straw polysaccharides for the production of reducing sugars. Carbohydrate Polymers, 71, 411–415.
Fang, H., Zhao, C., & Song, X. Y. (2010). Optimization of enzymatic hydrolysis of steam-exploded corn Stover by two approaches: Response surface methodology or using cellulase from mixed cultures of Trichoderma reesei RUT-C30 and Aspergillus niger NL02. Bioresource Technology, 101(11), 4111–4119.
Saratale, G. D., & Oh, S. E. (2011). Production of thermotolerant and alkalotolerant cellulolytic enzymes by isolated Nocardiopsis sp. KNU. Biodegradation, 22, 905–919.
Deswal, D., Khasa, Y. P., & Kuhad, R. C. (2011). Optimization of cellulase production by a brown rot fungus Fomitopsis sp. RCK2010 under solid state fermentation. Bioresource Technology, 102(10), 6065–6072.
Miller, G. L. (1959). Use of dinitrosalicylic acid reagent for determination of reducing sugar. Analytical Chemistry, 31, 426–428.
Ouyang, J., Li, Z., Li, X., Ying, H., & Yong, Q. (2009). Enhanced enzymatic conversion and glucose production via two-step enzymatic hydrolysis of corncob residue from xylo-oligosaccharides producers waste. Bioresources, 4, 1586–1599.
Kshirsagar, S. D., Bhalkar, B. N., Waghmare, P. R., Saratale, G. D., Saratale, R. G., & Govindwar, S. P. (2017). Sorghum husk biomass as a potential substrate for production of cellulolytic and xylanolytic enzymes by Nocardiopsis sp. KNU. 3 Biotech, 7, 163.
Saratale, G. D., Kshirsagar, S. D., Sampange, V. T., Saratale, R. G., Oh, S. E., Govindwar, S. P., & Oh, M. K. (2014). Cellulolytic enzymes production by utilizing agricultural wastes under solid state fermentation and its application for biohydrogen production. Applied Biochemistry and Biotechnology, 174(8), 2801–2817.
Saratale, G. D., Saratale, R. G., Lo, Y. C., & Chang, J. S. (2010). Multicomponent cellulase production by Cellulomonas biazotea NCIM-2550 and its applications for cellulosic biohydrogen production. Biotechnology Progress, 26, 406–416.
Kshirsagar, S., Saratale, G., Saratale, R., Govindwar, S., & Oh, M. K. (2016). An isolated Amycolatopsis sp. GDS for cellulase and xylanase production using agricultural waste biomass. Journal of Applied Microbiology, 120(1), 112–125.
Santos, T. C., Reis, N. S., Silva, T. P., Bonomo, R. C. F., Oliveira, E. A., Oliveira, J. R., & Franco, M. (2018). Production, optimisation and partial characterisation of enzymes from filamentous fungi using dried forage cactus pear as substrate. Waste and Biomass Valorization, 9, 571–579.
dos Santos, T. C., Abreu Filho, G., de Brito, A. R., Vieira Pires, A. J., Ferreira Bonomo, R. C., & Franco, M. (2016). Production and characterization of cellulolytic enzymes by Aspergillus niger and Rhizopus sp. by solid state fermentation of prickly pear. Revista Caatinga, 29, 222–233.
Oliveira, P., de Brito, A., Pimental, A., Soares, G., Pacheco, C., Santana, N., da Silva, E., Fernandes, A., Ferreira, M., de Oliveira, J., & Franco, M. (2019). Cocoa shell for the production of endoglucanase by Penicillium roqueforti ATCC 10110 in solid state fermentation and biochemical properties. Revista Mexicana De Ingeniería Química, 18, 777–787.
Ferraz, J. L. A. A., Souza, L. O., Fernandes, A. G. A., Oliveira, M. L. F., Oliveira, J. R., & Franco, M. (2020). Optimization of the solid-state fermentation conditions and characterization of xylanase produced by Penicillium roqueforti ATCC 10110 using yellow mombin residue (Spondias mombin L.). Chemical Engineering Communications, 207, 31–42.
Wei, H., Donohoe, B. S., Vinzant, T. B., Ciesielski, P. N., Wang, W., Gedvilas, L. M., Zeng, Y., Johnson, D. K., Ding, S. Y., Himmel, M. E., & Tucker, M. P. (2011). Elucidating the role of ferrous ion cocatalyst in enhancing dilute acid pretreatment of lignocellulosic biomass. Biotechnology for Biofuels, 4, 48.
Panagiotopoulos, I. A., Pasias, S., Bakker, R. R., de Vrije, T., Papayannakos, N., Claassen, P. A., & Koukios, E. G. (2013). Biodiesel and biohydrogen production from cotton-seed cake in a biorefinery concept. Bioresource Technology, 136, 78–86.
Walker, D. J., Ledesma, P., Delgado, O., & Breccia, J. (2006). High endo-β-1,4-D-glucanase activity in a broad pH range from the alkali tolerant Nocardiopsis sp. SES28. World Journal of Microbiology and Biotechnology, 22, 761–764.
Vasconcellos, V. M., Tardioli, P. W., Giordano, R. L. C., & Farinas, C. S. (2016). Addition of metal ions to a (hemi)cellulolytic enzymatic cocktail produced in-house improves its activity, thermostability, and efficiency in the saccharification of pretreated sugarcane bagasse. New Biotechnology, 33(3), 331–337.
Khalil, A. I. (2002). Production and characterization of cellulolytic and xylanolytic enzymes from the ligninolytic white-rot fungus Phanerochaete chrysosporium grown on sugarcane bagasse. World Journal of Microbiology and Biotechnology, 18, 753–759.
Soni, R., Sandhu, D. K., & Soni, S. K. (1999). Localisation and optimisation of cellulase production in Chaetomium erraticum. Journal of Biotechnology, 73, 43–51.
Romero, M. D., Aguado, J., Gonzalez, L., & Ladero, M. (1999). Cellulase production by Neurospora crassa on wheat straw. Enzyme and Microbial Technology, 25, 244–250.
Guevara, C., & Zambrano, M. M. (2006). Sugarcane cellulose utilization by a defined microbial consortium. FEMS Microbiology Letters, 255(1), 52–58.
Tang, X., He, L. Y., Tao, X. Q., Dang, Z., Guo, C. L., Lu, G. N., & Yi, X. Y. (2010). Construction of an artificial microalgal-bacterial consortium that efficiently degrades crude oil. Journal of Hazardous Materials, 181(1-3), 1158–1162.
Lin, H., Wang, B., Zhuang, R. Y., Zhou, Q. F., & Zhao, Y. H. (2011). Artificial construction and characterization of a fungal consortium that produces cellulolytic enzyme system with strong wheat straw saccharification. Bioresource Technology, 102(22), 10569–10576.
Kausar, H., Sariah, M., Saud, H. M., Alam, M. Z., & Ismail, M. R. (2010). Development of compatible lignocellulolytic fungal consortium for rapid composting of rice straw. International Biodeterioration and Biodegradation, 64, 594–600.
Ahamed, A., & Vermette, P. (2008). Enhanced enzyme production from mixed cultures of Trichoderma reesei RUT-C30 and Aspergillus niger LMA grown as fed batch in a stirred tank bioreactor. Biochemical Engineering Journal, 42, 41–46.
Kolasa, M., Ahring, B. K., Lubeck, P. S., & Lubeck, M. (2014). Co-cultivation of Trichoderma reesei RutC30 with three black Aspergillus strains facilitates efficient hydrolysis of pretreated wheat straw and shows promises for on-site enzyme production. Bioresource Technology, 169, 143–148.
Cherry, J. R., & Fidantsef, A. L. (2003). Directed evolution of industrial enzymes: An update. Current Opinion in Biotechnology, 14(4), 438–443.
Szijarto, N., Faigl, Z., Reczey, K., Mezes, M., & Bersenyi, A. (2004). Cellulase fermentation on a novel substrate (waste cardboard) and subsequent utilization of home-produced cellulase and commercial amylase in a rabbit feeding trial. Industrial Crops and Products, 20, 49–57.
Fierobe, H. P., Mingardon, F., Mechaly, A., Belaich, A., Rincon, M. T., Pages, S., Lamed, R., Tardif, C., Belaich, J. P., & Bayer, E. A. (2005). Action of designer cellulosomes on homogeneous versus complex substrates - controlled incorporation of three distinct enzymes into a defined trifunctional scaffoldin. Journal of Biological Chemistry, 280(16), 16325–16334.
Sukumaran, R. K., Singhania, R. R., & Pandey, A. (2005). Microbial cellulases-production, applications and challenges. Journal of Science and Industrial Research, 64, 832–844.
Vanee, N., Brooks, J. P., & Fong, S. S. (2017). Metabolic profile of the cellulolytic industrial Actinomycete Thermobifida fusca. Metabolites, 7, 57.
Kumar, A., Gautam, A., & Dutt, D. (2016). Co-cultivation of Penicillium sp. AKB-24 and Aspergillus nidulans AKB-25 as a cost-effective method to produce cellulases for the hydrolysis of pearl millet stover. Fermentation, 2, 12.
Zhang, Y. H. P., & Lynd, L. R. (2006). A functionally based model for hydrolysis of cellulose by fungal cellulase. Biotechnology and Bioengineering, 94(5), 888–898.
Visser, E. M., Falkoski, D. L., de Almeida, M. N., Maitan-Alfenas, G. P., & Guimaraes, V. M. (2013). Production and application of an enzyme blend from Chrysoporthe cubensis and Penicillium pinophilum with potential for hydrolysis of sugarcane bagasse. Bioresource Technology, 144, 587–594.
Adsul, M., Sharma, B., Singhania, R. R., Saini, J. K., Sharma, A., Mathur, A., Gupta, R., & Tuli, D. K. (2014). Blending of cellulolytic enzyme preparations from different fungal sources for improved cellulose hydrolysis by increasing synergism. RSC Advances, 4, 44726–44732.
Saini, J. K., Singhania, R. R., Satlewal, A., Saini, R., Gupta, R., Tuli, D., Mathur, A., & Adsul, M. (2016). Improvement of wheat straw hydrolysis by cellulolytic blends of two Penicillium sp. Renewable Energy, 98, 43–50.
Hu, J. G., Arantes, V., & Saddler, J. N. (2011). The enhancement of enzymatic hydrolysis of lignocellulosic substrates by the addition of accessory enzymes such as xylanase: Is it an additive or synergistic effect? Biotechnology for Biofuels, 4, 36.
Acknowledgments
The authors would like to acknowledge authorities of Shivaji University, Kolhapur, India.
Funding
This research work was supported by Hanyang University (HY201800000003220) to SPG. MBK and BHJ thank National Research Foundation of Korea (NRF), Ministry of Education, Science, and Technology (MEST) for the grant (NRF-2017R1A2B2004143).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflicts of interest.
Research Involving Human Participants and/or Animals
No involvement of human participants and animals.
Informed Consent
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Fig. S1
(DOCX 58 kb).
Rights and permissions
About this article
Cite this article
Kshirsagar, S., Waghmare, P., Saratale, G. et al. Composition of Synthesized Cellulolytic Enzymes Varied with the Usage of Agricultural Substrates and Microorganisms. Appl Biochem Biotechnol 191, 1695–1710 (2020). https://doi.org/10.1007/s12010-020-03297-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-020-03297-8