Abstract
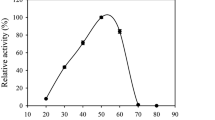
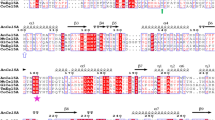
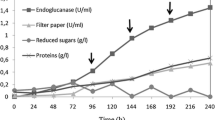
Enzyme reaction products and by-products from pretreatment steps can inhibit endoglucanases and are major factors limiting the efficiency of enzymatic lignocellulosic biomass hydrolysis. The gene encoding the endoglucanase from Scytalidium thermophilum (egst) was cloned and expressed as a soluble protein in Pichia pastoris GS115. The recombinant enzyme (Egst) was monomeric (66 kDa) and showed an estimated carbohydrate content of 53.3% (w/w). The optimum temperature and pH of catalysis were 60–70 °C and pH of 5.5, respectively. The enzyme was highly stable at pH 3.0–8.0 with a half-life in water of 100 min at 65 °C. The Egst presented good halotolerance, retaining 84.1 and 71.4% of the control activity in the presence of 0.5 and 2.0 mol L−1 NaCl, respectively. Hydrolysis of medium viscosity carboxymethylcellulose (CMC) by Egst was stimulated 1.77-, 1.84-, 1.64-, and 1.8-fold by dithiothreitol, β-mercaptoethanol, cysteine, and manganese at 10, 10, 10, and 5 mmol L−1 concentration, respectively. The enzyme hydrolyzed CMC with maximal velocity and an apparent affinity constant of 432.10 ± 16.76 and 10.5 ± 2.53 mg mL−1, respectively. Furthermore, the Egst was tolerant to reaction products and able to act on pretreated fractions sugarcane bagasse demonstrating excellent properties for application in the hydrolysis of lignocellulosic biomass.






Similar content being viewed by others
References
Meleiro, L. P., Salgado, J. C. S., Maldonado, R. F., Alponti, J. S., Zimbardi, A. L. R. L., Jorge, J. A., Ward, R. J., & Furriel, R. P. M. (2015). A Neurospora crassa β-glucosidase with potential for lignocellulose hydrolysis shows strong glucose tolerance and stimulation by glucose and xylose. Journal of Molecular Catalysis B: Enzymatic, 122, 131–140. https://doi.org/10.1016/j.molcatb.2015.09.003.
Arevalo-Gallegos, A., Ahmad, Z., Asgher, M., Parra-Saldivar, R., & Iqbal, H. M. N. (2017). Lignocellulose: A sustainable material to produce value-added products with a zero waste approach—A review. International Journal of Biological Macromolecules, 99, 308–318. https://doi.org/10.1016/j.ijbiomac.2017.02.097.
Teugjas, H., & Väljamäe, P. (2013). Selecting β-glucosidases to support cellulases in cellulose saccharification. Biotechnology for Biofuels, 6(1), 105. https://doi.org/10.1186/1754-6834-6-105.
De Souza, A. P., Leite, D. C. C., Pattathil, S., Hahn, M. G., & Buckeridge, M. S. (2013). Composition and structure of sugarcane cell wall polysaccharides: Implications for second-generation bioethanol production. Bioenergy Research, 6, 64–579.
Boisset, C., Fraschini, C., Schülein, M., Henrissat, B., & Chanzy, H. (2000). Imaging the enzymatic digestion of bacterial cellulose ribbons reveals the endo character of the cellobiohydrolase Cel6A from Humicola insolens and its mode of synergy with cellobiohydrolase Cel7A. Applied and Environmental Microbiology, 66(4), 1444–1452. https://doi.org/10.1128/AEM.66.4.1444-1452.2000.
Yennamalli, R. M., Rader, A. J., Kenny, A. J., Wolt, J. D., & Sen, T. Z. (2013). Endoglucanases: insights into thermostability for biofuel applications. Biotechnology for Biofuel, 6(1), 136. https://doi.org/10.1186/1754-6834-6-136.
Sorensen, A., Lübeck, M., Lübeck, P. S., & Ahring, B. K. (2013). Fungal beta-glucosidases: a bottleneck in industrial use of lignocellulosic materials. Biomolecules, 3(3), 612–631. https://doi.org/10.3390/biom3030612.
Loaces, I., Bottini, G., Moyna, G., Fabiano, E., Martínez, A., & Noya, F. (2016). EndoG: a novel multifunctional halotolerant glucanase and xylanase isolated from cow rumen. Journal of Molecular Catalysis B: Enzymatic, 126, 1–9. https://doi.org/10.1016/j.molcatb.2016.01.004.
Carli, S., Meleiro, L. P., Rosa, J. C., Moraes, L. A. B., Jorge, J. A., Masui, D. C., & Furriel, R. P. M. (2017). A novel thermostable and halotolerant xylanase from Colletotrichum graminicola. Journal of Molecular Catalaysis B: Enzymatic. https://doi.org/10.1016/j.molcatb.2017.05.002.
Xue, D., Lin, D., Gong, C., Peng, C., & Yao, S. (2017). Expression of a bifunctional cellulase with exoglucanase and endoglucanase activities to enhance the hydrolysis ability of cellulase from a marine Aspergillus niger. Process Biochemistry, 52, 115–122. https://doi.org/10.1016/j.procbio.2016.09.030.
Silva, J. C. R., Guimarães, L. H. S., Salgado, J. C. S., Furriel, R. P. M., Polizeli, M. L. T. M., Rosa, J. C., & Jorge, J. A. (2013). Purification and biochemical characterization of glucose–cellobiose-tolerant cellulases from Scytalidium thermophilum. Folia Microbiologica, 58(6), 561–568. https://doi.org/10.1007/s12223-013-0245-7.
Temelli, F. (1997). Extraction and functional properties of barley β-glucan as affected by temperature and pH. Journal of Food Science, 62(6), 1194–1201. https://doi.org/10.1111/j.1365-2621.1997.tb12242.x.
Miller, G. L. (1959). Use of dinitrosalicylic acid reagent for determination of reducing sugar. Analytical Chemistry, 31(3), 426–428. https://doi.org/10.1021/ac60147a030.
Souza, F. H. M., Nascimento, C. V., Rosa, J. C., Masui, D. C., Leone, F. A., Jorge, J. A., & Furriel, R. P. M. (2010). Purification and biochemical characterization of a mycelial glucose- and xylose-stimulated β-glucosidase from the thermophilic fungus Humicola insolens. Process Biochemistry, 45(2), 272–278. https://doi.org/10.1016/j.procbio.2009.09.018.
Read, S. M., & Northcote, D. H. (1981). Minimization of variation in the response to different proteins of the Coomassie blue G dye-binding assay for protein. Analytical Biochemistry, 116(1), 53–64. https://doi.org/10.1016/0003-2697(81)90321-3.
Dubois, M., Gilles, K. A., Hamilton, J. K., Rebers, P. A., & Smith, F. (1956). Colorimetric method for determination of sugars and related substances. Analytical Chemistry, 28(3), 350–356. https://doi.org/10.1021/ac60111a017.
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227(5259), 680–685. https://doi.org/10.1038/227680a0.
McIlvaine, T. C. (1921). A buffer solution for colorimetric comparison. The Journal of Biological Chemistry, 49, 183–186.
Leone, F. A., Baranauskas, J. A., Furriel, R. P. M., & Borin, I. A. (2005). SigrafW: an easy-to-use program for fitting enzyme kinetic data. Biochemistry and Molecular Biology Education, 33(6), 399–403. https://doi.org/10.1002/bmb.2005.49403306399.
Segel, I. H. (1975). Enzyme kinetics: behavior and analysis of rapid equilibrium and steady-state enzyme systems. New York: Wiley-Interscience.
Rowley, J., Decker, S. R., Michener, W., & Black, S. (2013). Efficient extraction of xylan from delignified corn stover using dimethyl sulfoxide. 3. Biotech, 3, 433–438.
Henrissat, B., Callebaut, I., Fabrega, S., Lehn, P., Mornon, J. P., & Davies, G. (1995). Conserved catalytic machinery and the prediction of a common fold for several families of glycosyl hydrolases. Proceedings of the National Academy of Sciences, 92(15), 7090–7094. https://doi.org/10.1073/pnas.92.15.7090.
Rubini, M. R., Dillon, A. J. P., Kyaw, C. M., Faria, F. P., Poças-Fonseca, M. J., & Silva-Pereira, I. (2010). Cloning, characterization and heterologous expression of the first Penicillium echinulatum cellulase gene. Journal of Applied Microbiology, 108(4), 1187–1198. https://doi.org/10.1111/j.1365-2672.2009.04528.x.
Zhao, X., Zhang, L., & Liu, D. (2012). Biomass recalcitrance. Part I: the chemical compositions and physical structures affecting the enzymatic hydrolysis of lignocellulose. Biofuels Bioproduct and Biorefining, 6(4), 465–482. https://doi.org/10.1002/bbb.1331.
Liu, G., Qin, Y., Hu, Y., Gao, M., Peng, S., & Qu, Y. (2013). An endo-1,4-β-glucanase PdCel5C from cellulolytic fungus Penicillium decumbens with distinctive domain composition and hydrolysis product profile. Enzyme and Microbial Technology, 52, 190–195.
Li, X., & Yu, H. Y. (2012). Purification and characterization of an organic-solvent-tolerant cellulase from a halotolerant isolate, Bacillus sp. L1. Journal of Industrial Microbiology & Biotechnology, 39(8), 1117–1124. https://doi.org/10.1007/s10295-012-1120-2.
Liu, J., Liu, W., Zhao, X., Shen, W., Cao, H., & Cui, Z. (2011). Cloning and functional characterization of a novel endo-β-1,4-glucanase gene from a soil-derived metagenomic library. Applied Microbiology and Biotechnology, 89(4), 1083–1092. https://doi.org/10.1007/s00253-010-2828-4.
Karnaouri, A., Muraleedharan, M. N., Dimarogona, M., Topakas, E., Rova, U., Sandgren, M., & Christakopoulos, P. (2017). Recombinant expression of thermostable processive MtEG5 endoglucanase and its synergism with MtLPMO from Myceliophthora thermophila during the hydrolysis of lignocellulosic substrates. Biotechnology for Biofuels, 10(1), 126. https://doi.org/10.1186/s13068-017-0813-1.
Xiong, W., Yang, J. K., Chen, F. Y., & Han, Z. G. (2017). The catalytic domain of Penicillium crustosum endoglucanase EGL1 has cellulose-binding capacity and cellulolytic activity. Enzyme and Microbial Technology, 97, 71–81.
Phadtare, P., Joshi, S., & Satyanarayana, T. (2017). Recombinant thermo-alkali-stable endoglucanase of Myceliopthora thermophila BJA (rMt-egl): biochemical characteristics and applicability in enzymatic saccharification of agro-residues. International Journal of Biological Macromolecules, 104(Pt A), 107–116. https://doi.org/10.1016/j.ijbiomac.2017.05.167.
Huy, N. D., Nguyen, C. L., Park, H. S., Loc, N. H., Choi, M. S., Kim, D. H., Seo, J. W., & Park, S. M. (2016). Characterization of a novel manganese dependent endoglucanase belongs in GH family 5 from Phanerochaete chrysosporium. Journal of Bioscience and Bioengineering, 121(2), 154–159. https://doi.org/10.1016/j.jbiosc.2015.06.009.
Pan, R., Hu, Y., Long, L., Wang, J., & Ding, S. (2016). Extra carbohydrate binding module contributes to the processivity and catalytic activity of a non-modular hydrolase family 5 endoglucanase from Fomitiporia mediterranea MF3/22. Enzyme and Microbial Technology, 91, 42–51.
Liu, G., Li, Q., Shang, N., Huang, J. W., Ko, T. P., Liu, W., Zheng, Y., Han, X., Chen, Y., Chen, C. C., Jinf, J., & Guo, R. T. (2016). Functional and structural analyses of a 1,4-β-endoglucanase from Ganoderma lucidum. Enzyme and Microbial Technology, 86, 67–74.
Jin, X., Meng, N., & Xia, L. (2011). Expression of an Endo-β-1,4-glucanase Gene from Orpinomyces PC-2 in Pichia pastoris. International Journal of Molecular Sciences, 12(12), 3366–3380. https://doi.org/10.3390/ijms12053366.
Azhar, S. H. M., Abdulla, R., Jambo, S. A., Marbawi, H., Gansau, J. A., Faik, A. A. M., & Rodrigues, K. F. (2017). Yeasts in sustainable bioethanol production: A review. Biochemistry and Biophysics Report, 10, 52–61.
Dave, B. R., Sudhir, A. P., & Subramanian, R. B. (2015). Purification and properties of an endoglucanase from Thermoascus aurantiacus. Biotechnol Reports, 6, 85–90. https://doi.org/10.1016/j.btre.2014.11.004.
Kaur, J., Chadha, B. S., Kumar, B. A., & Saini, H. S. (2007). Purification and characterization of two endoglucanases from Melanocarpus sp. MTCC 3922. Bioresource Technology, 98(1), 74–81. https://doi.org/10.1016/j.biortech.2005.11.019.
Saha, B. C. (2004). Production, purification and properties of endoglucanase from a newly isolated strain of Mucor circinelloides. Process Biochemistry, 39(12), 1871–1876. https://doi.org/10.1016/j.procbio.2003.09.013.
de María, P. D. (2013). On the use of seawater as reaction media for large-scale applications in biorefineries. Chem Cat Chem, 00, 1–7.
Benhmad, I., Boudabbous, M., Yaich, A., Rebai, M., & Gargouri, A. (2016). A novel neutral, halophile Stachybotrys microspora-based endoglucanase active impact on β-glucan. Bioprocessand Biosystem Engineering, 39, 685–693.
Cheng, R., Xu, L., Wang, S., Wang, Y., & Zhang, J. (2014). Recombinant expression and characterization of an acid-, alkali- and salt-tolerant β-1,3-1,4-glucanase from Paenibacillus sp. S09. Biotechnology Letters, 36(4), 797–803. https://doi.org/10.1007/s10529-013-1413-1.
Dong, M., Yang, Y., Tang, X., Shen, J., Xu, B., Li, J., Wu, Q., Zhou, J., Ding, J., Han, N., Mu, Y., & Huang, Z. (2016). NaCl-, protease-tolerant and cold-active endoglucanase from Paenibacillus sp. YD236 isolated from the feces of Bos frontalis. SpringerPlus, 5(1), 746. https://doi.org/10.1186/s40064-016-2360-9.
Hmad, I. B., Boudabbous, M., Belghith, H., & Gargouri, A. (2017). A novel ionic liquid-stable halophilic endoglucanase from Stachybotrys microspora. Process Biochemistry, 54, 59–66. https://doi.org/10.1016/j.procbio.2017.01.007.
Lee, J. M., Moon, S. Y., Kim, Y. R., Kim, K. W., Lee, B. J., & Kong, I. S. (2017). Improvement of thermostability and halostability of β-1,3-1,4-glucanase by substituting hydrophobic residue for Lys 48. International Journal of Biological Macromolecules, 94(Pt A), 594–602. https://doi.org/10.1016/j.ijbiomac.2016.10.043.
Garg, R., Srivastava, R., Brahma, V., Verma, L., Karthikeyan, S., & Sahni, G. (2016). Biochemical and structural characterization of a novel halotolerant cellulase from soil metagenome. Scientific Reports, 23(6), 39634.
Zhu, C., Xu, Z., & Song, R. (2011). The endoglucanase from Bacillus subtilis BEC-1 bears halo-tolerant, acidophilic and dithiothreitol-stimulated enzyme activity. World Journal of Microbiology and Biotechnology, 27(12), 2863–2871. https://doi.org/10.1007/s11274-011-0767-6.
Siddiqui, K. S., Azhar, M. J., Rashid, M. H., Ghuri, T. M., & Rajoka, M. I. (1997). Purification and the effect of manganese ions on the activity of carboxymethylcellulases from Aspergillus niger and Cellulomonas biazotea. Folia Microbiologica, 43, 303–311.
Tao, Y. M., Zhu, X. Z., Huang, J. Z., Ma, S. J., Wu, X. B., Long, M. N., & Chen, Q. X. (2010). Purification and properties of endoglucanase from a sugar cane bagasse hydrolyzing strain, Aspergillus glaucus XC9. Journal of Agricultural and Food Chemistry, 58(10), 6126–6130. https://doi.org/10.1021/jf1003896.
Jabbar, A., Rashid, M. H., Javed, M. R., Perveen, R., & Malana, M. A. (2008). Kinetics and thermodynamics of a novel endoglucanase (CMCase) from Gymnoascella citrina produced under solid-state condition. Journal of Industrial Microbiology, 35(6), 515–524. https://doi.org/10.1007/s10295-008-0310-4.
Nakamura, K., & Kitamura, K. (1983). Purification and some properties of an alkaline xylanase from alkaliphilic Bacillus sp. Strain 41M-1. Journal of Fermentation Technology, 61, 379–382.
Wang, J., Kang, L., Liu, Z., & Yuan, S. (2017). Gene cloning, heterologous expression and characterization of a Coprinopsis cinerea endo-β-1,3(4)-glucanase. Fungal Biology, 121(1), 61–68. https://doi.org/10.1016/j.funbio.2016.09.003.
Acknowledgements
We thank André Justino for technical assistance.
Funding
This investigation was supported by research grants from CNPq (Conselho deDesenvolvimento Científico e Tecnológico), FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo) and CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior). Luana P. Meleiro and Raquel F. Maldonado received post-doctoral scholarship from FAPESP; Marcela S. Torricillas received a scientific initiation scholarship from CNPq; Sibeli Carli received a Ph.D. scholarship from CAPES; Richard J. Ward; João A. Jorge and Rosa P. M. Furriel received researcher stipends from CNPq.
Author information
Authors and Affiliations
Contributions
Luana P. Meleiro, Sibeli Carli and Rosa P. M. Furriel conceived and designed the experiments; Luana P. Meleiro, Sibeli Carli, Raquel F. Maldonado, Marcela S. Torricillas and Ana L. R. L. Zimbardi performed the experiments; Raquel F. Maldonado and Richard J. Ward conducted the cloning and expression experiments; Luana P. Meleiro, Sibeli Carli and Marcela S. Torricillas analyzed the data; Richard J. Ward, Rosa P. M. Furriel and João A. Jorge contributed reagents, materials and analysis tools; Luana P. Meleiro and Sibeli Carli wrote the paper.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Meleiro, L.P., Carli, S., Fonseca-Maldonado, R. et al. Overexpression of a Cellobiose-Glucose-Halotolerant Endoglucanase from Scytalidium thermophilum . Appl Biochem Biotechnol 185, 316–333 (2018). https://doi.org/10.1007/s12010-017-2660-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-017-2660-8