Abstract
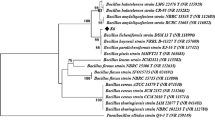
Paenibacillus sp. S29 was isolated from soil and produces an alginate lyase. The molecular weight of this enzyme was 32 kDa and the N-terminal amino acid sequence was ASVTKST. The optimal pH was approximately 8.7 and the enzyme was stable over a pH range of 5.6 to 8.8 at 40 °C for 60 min. The optimal temperature was approximately 50 °C, and the residual activity was not decreased at temperatures of up to 40 °C at pH 8 for 30 min. Paenibacillus sp. S29 alginate lyase had also a little activity toward hyaluronic acid. Poly G and poly M separated from alginate were degraded efficiently, and poly M was the more susceptible substrate. The maximum amount of reducing sugar released by the enzyme was 261 mg per gram of sodium alginate. The main sugar released was monosaccharide (unsaturated uronate) and small amounts of oligosaccharides of degree of polymerization 2–6 were also released.






Similar content being viewed by others
References
Wong, T. Y., Preston, L. A., & Schiller, N. L. (2000). Alginate lyase: review of major sources and enzyme characteristics, structure-function analysis, biological roles, and applications. Annual Review of Microbiology, 54, 289–340.
Akiyama, H., Endo, T., Nakakita, R., Murata, K., Yonemoto, Y., & Okayama, K. (1992). Effect of depolymerized alginates on the growth of Bifidobacteria. Bioscience, Biotechnology, and Biochemistry, 56, 355–356.
Kobayashi, N., Kanazawa, Y., Yamabe, S., Iwata, K., Nishizawa, M., Yamagishi, T., Nishikaze, O., & Tsuji, K. (1997). Effects of depolymerized sodium alginate on serum total cholesterol in healthy women with a high cholesterol intake. Journal of Home Economics of Japan, 48, 225–230.
Zhang, Z., Yu, G., Guan, H., Zhao, X., Du, Y., & Jiang, X. (2004). Preparation and structure elucidation of alginate oligosaccharides degraded by alginate lyase from Vibrio sp. 510. Carbohydrate Research, 339, 1475–1481.
Zhu, B., Tana, H., Qin, Y., Xu, Q., Du, Y., & Yin, H. (2015). Characterization of a new endo-type alginate lyase from Vibrio sp. W13. International Journal of Biological Macromolecules, 75, 330–337.
Natsume, M., Kamo, Y., Hirayama, M., & Adachi, T. (1994). Isolation and characterization of alginate-derived oligosaccharides with root growth-promoting activities. Carbohydrate Research, 258, 187–197.
Falkeborg, M., Cheong, L. Z., Gianfico, C., Sztukiel, K. M., Kristensen, K., Glasius, M., Xu, X., & Guo, Z. (2014). Alginate oligosaccharides: enzymatic preparation and antioxidant property evaluation. Food Chemistry, 164, 185–194.
Jagtap, S. S., Hehemann, J. H., Polz, M. F., Lee, J. K., & Zhao, H. (2014). Comparative biochemical characterization of three exolytic oligoalginate lyases from Vibrio splendidus reveals complementary substrate scope, temperature, and pH adaptations. Applied and Environmental Microbiology, 80, 4207–4214.
Kim, H. T., Chung, J. H., Wang, D., Lee, J., Woo, H. C., Choi, I. G., & Kim, K. H. (2012). Depolymerization of alginate into a monomeric sugar acid using Alg17C, an exo-oligoalginate lyase cloned from Saccharophagus degradans 2-40. Applied Microbiology and Biotechnology, 93, 2233–2239.
Takase, R., Ochiai, A., Mikami, B., Hashimoto, W., & Murata, K. (2010). Molecular identification of unsaturated uronate reductase prerequisite for alginate metabolism in Sphingomonas sp. A1. Biochimica et Biophysica Acta, General Subjects, 1804, 1925–1936.
Lee, O. K., & Lee, E. Y. (2016). Sustainable production of bioethanol from renewable brown algae biomass. Biomass and Bioenergy, 92, 70–75.
Gasesa, P., & Wusteman, F. S. (1990). Plate assay for simultaneous detection of alginate lyases and determination of substrate specificity. Applied and Environmental Microbiology, 56, 2265–2267.
Miller, G. L. (1959). Use of dinitrosalicilic acid reagent for determination of reducing sugar. Analytical Chemistry, 31, 426–428.
Iwamoto, Y., Araki, R., Iriyama, K., Oda, T., Fukuda, H., Hayashida, S., & Muramatsu, T. Purification and characterization of bifunctional alginate lyase from Alteromonas sp. strain no. 272 and its action on saturated oligomeric substrates. Bioscience, Biotechnology, and Biochemistry, 65, 133–142.
Li, J. W., Dong, S., Song, J., Li, C. B., Chen, X. L., Xie, B. B., & Zhang, Y. Z. (2011). Purification and characterization of a bifunctional alginate lyase from Pseudoalteromonas sp. SM0524. Marine Drugs, 9, 109–123.
Inoue, A., Takadono, K., Nishiyama, R., Tajima, K., Kobayashi, T., & Ojima, T. (2014). Characterization of an alginate lyase, FlAlyA, from Flavobacterium sp. strain UMI-01 and its expression in Escherichia coli. Marine Drugs, 12, 4693–4712.
Swift, S M. Hudgens, J. W. Heselpoth, R. D. Bales, P. M., & Nelson, D. C. (2014). Characterization of AlgMsp, an alginate lyase from Microbulbifer sp. 6532A. PLOS one, November 19.
Lee, S. I., Choi, S. H., Lee, E. Y., & Kim, H. S. (2012). Molecular cloning, purification, and characterization of a novel polyMG-specific alginate lyase responsible for alginate MG block degradation in Stenotrophomas maltophilia KJ-2. Applied Microbiology and Biotechnology, 95, 1643–1653.
Yamasaki, M., Moriwaki, S., Miyake, O., Hashimoto, W., Murata, K., & Mikami, B. (2004). It exhibited preferential β-elimination activity towards polyMG-block (100%) and then followed by polyM-block (16.8%) and polyG-block(1.83%). The Journal of Biological Chemistry, 279, 31863–31872.
Badur, A. H., Jagtap, S. S., Yalamanchili, G., Lee, J. K., Zhao, H., & Rao, C. V. (2015). Characterization of the alginate lyases from Vibrio splendidus 12B01 are endolytic. Applied and Environmental Microbiology, 81, 1865–1873.
Huang, L., Zhou, J., Li, X., Peng, Q., Lu, H., & Du, Y. (2013). Characterization of a new alginate lyase from newly isolated Flavobacterium sp. S20. Journal of Industrial Microbiology & Biotechnology, 40, 113–122.
Wang, D. M., Kim, H. T., Yun, E. J., Kim, D. H., Park, Y., Woo, H. C., & Kim, K. H. (2014). Optimal production of 4-deoxy-L-erythro-5-hexoseulose uronic acid from alginate for brown macro algae saccharification by combining endo- and exo-type alginate lyases. Bioprocess and Biosystems Engineering, 37, 2105–2111.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kurakake, M., Kitagawa, Y., Okazaki, A. et al. Enzymatic Properties of Alginate Lyase from Paenibacillus sp. S29. Appl Biochem Biotechnol 183, 1455–1464 (2017). https://doi.org/10.1007/s12010-017-2513-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-017-2513-5