Abstract
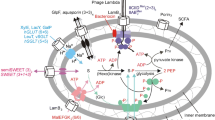
Malonyl-CoA plays an important role in the synthesis and elongation of fatty acids in yeast Saccharomyces cerevisiae. Malonyl-CoA is at a low concentration inside the cell and is produced mainly from acetyl-CoA through the enzyme acetyl-CoA carboxylase. It would be beneficial to find an alternative source of malonyl-CoA to increase its intracellular concentration and overall synthesis of the fatty acids. MatB gene from the bacteria Rhizobium leguminosarium bv. trifolii encodes for a malonyl-CoA synthetase which catalyzes the formation of the malonyl-CoA directly from malonate and CoA. However, results from high-performance liquid chromatography (HPLC) proved that Saccharomyces cerevisiae itself does not contain enough cytoplasmic malonate within them and is unable to uptake exogenously supplied malonate in the form of malonic acid. A dicarboxylic acid plasma membrane transporter with the ability to uptake exogenous malonic acid was identified from another species of yeast known as Schizosaccharomyces pombe and the gene encoding this transporter is identified as the mae1 gene. From the experiments thus far, the mae1 gene had been successfully cloned and transformed into Saccharomyces cerevisiae. The expression and functional ability of the encoded plasma membrane dicarboxylic acid transporter were also demonstrated and verified using specialized technologies such as RT-PCR, yeast immunofluorescence, HPLC, and LC-MS.









Similar content being viewed by others
References
Energy Information Administration (2007) International Energy Outlook 2007, US Department of Energy. http://www.eia.doe.gov/oiaf/ieo/pdf/0484.(2007).pdf. Accessed Jan 2011.
Demirbas, A. (2009). Political, economic and environmental impacts of biofuels: a review. Applied Energy, 86(1), S108–S117.
Fortman, J. L., et al. (2008). Biofuel alternatives to ethanol: pumping the microbial well. Trends in Biotechnology, 26(7), 375–381.
Ejsing, C. S., et al. (2009). Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proceedings of the National Academy of Sciences of the United States of America, 106(7), 2136–2141.
Tehlivets, O., Scheuringer, K., & Kohlwein, S. (2007). Fatty acid synthesis and elongation in yeast. Bba-Mol Cell Biol Lipids, 177(3), 255–270.
Kim, Y. S. (2002). Malonate metabolism: biochemistry, molecular biology, physiology, and industrial application. Journal of Biochemistry and Molecular Biology, 35(5), 443–451.
Casal, M., et al. (2008). Transport of carboxylic acids in yeasts. FEMS Microbiology Reviews, 32(6), 974–994.
Camarasa, C., et al. (2001). Characterization of Schizosaccharomyces pombe malate permease by expression in Saccharomyces cerevisiae. Applied and Environmental Microbiology, 67(9), 4144–4151.
RESTEK, analysis of preservatives using HPLC, in HPLC application note #59398.
Ross, P. L., et al. (2004). Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Molecular & cellular proteomics : MCP, 3(12), 1154–1169.
Hansen, K. C., et al. (2003). Mass spectrometric analysis of protein mixtures at low levels using cleavable 13c-isotope-coded affinity tag and multidimensional chromatography. Molecular & Cellular Proteomics, 2(5), 299–314.
Zappacosta, F., & Annan, R. S. (2004). N-terminal isotope tagging strategy for quantitative proteomics: results-driven analysis of protein abundance changes. Analytical Chemistry, 76(22), 6618–6627.
Sui, J., et al. (2008). Comparative proteomics analysis of vascular smooth muscle cells incubated with S- and R-enantiomers of atenolol using iTRAQ-coupled two-dimensional LC-MS/MS. Molecular & cellular proteomics : MCP, 7(6), 1007–1018.
Greene, J. G., et al. (1993). Inhibition of succinate dehydrogenase by malonic acid produces an "excitotoxic" lesion in rat striatum. Journal of Neurochemistry, 61(3), 1151–1154.
Kondrashov, F. A., et al. (2006). Evolution of glyoxylate cycle enzymes in Metazoa: evidence of multiple horizontal transfer events and pseudogene formation. Biology Direct, 1, 31.
Samokhvalov, V., Ignatov, V., & Kondrashova, M. (2004). Inhibition of Krebs cycle and activation of glyoxylate cycle in the course of chronological aging of Saccharomyces cerevisiae. Compensatory role of succinate oxidation. Biochimie, 86(1), 39–46.
Lorenz, M. C., & Fink, G. R. (2002). Life and death in a macrophage: role of the glyoxylate cycle in virulence. Eukaryotic Cell, 1(5), 657–662.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chen, W.N., Tan, K.Y. “Malonate Uptake and Metabolism in Saccharomyces cerevisiae”. Appl Biochem Biotechnol 171, 44–62 (2013). https://doi.org/10.1007/s12010-013-0334-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-013-0334-8