Abstract
The study analysed mechanically deboned chicken meat subjected to washing and separation of fat and connective tissue in order to produce a preparation of myofibrillar proteins (MP). The preparation was modified using microbial transglutaminase (MTG). For this purpose, MP was supplemented with 3 g/kg MTG and modified at 6–7 °C for 5 h. Changes in the dynamics of water binding, which occurred in the course of heating and cooling within the temperature range of 20–70–20 °C, were analysed in the tested systems. Relaxation times T 1 and T 2 were determined using the low-field nuclear magnetic resonance (NMR). Enzymatic modification of MP and heating resulted in significant changes in the dynamics of bulk water binding in the tested protein systems. Due to the presence of transglutaminase in the preparation, the changes begin and end at lower temperatures. Analysis of relaxation time T 1 showed that the addition of the enzyme to MP causes water binding in the sample up to 48 h after gel cooling. The addition of the enzyme to MP reduces the energy barrier determined, based on activation energy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The considerable amount of meat left on poultry carcasses after trimming of breast and thigh muscles has been an incentive for the development of effective methods for its recovery. Material obtained as a result of mechanical deboning (mechanically recovered meat, MRM) is characterised by a variable chemical composition and low microbiological stability as well as a high variability of functional processing traits (Trindade et al. 2004; Negrão et al. 2005; Serdaroğlu and Turp 2005; EFSA 2013). The composition of proteins in poultry MRM indicates that their most valuable fraction is composed of myofibrillar proteins. No other fraction of muscle proteins and no other protein substitute used in the food industry exhibit such advantageous functional properties as myofibrillar proteins. The results of many studies clearly indicated that based on the principles of fish surimi technology, it is feasible and viable to recover from mechanically recovered poultry meat (MRPM) this technologically and nutritionally most valuable protein fraction (Yang and Froning 1994; Nowsad et al. 2000; Stangierski and Kijowski 2000; Perlo et al. 2006; Ramadhan et al. 2011; Cortez-Vega et al. 2012, 2015). The production method of fish surimi was developed and patented in Japan in the 1960s. Surimi is an original Japanese term referring to white, odourless and tasteless myofibril preparations obtained from mechanically recovered fish meat, washed several times with water and mixed with cryoprotectants (Lee 1984). European countries expressed their interest in this technology already in the early 1970s. Fractionation of MRPM may be divided into three stages: mixing of the raw material with water, buffer solution or low ionic strength aqueous saline solution; separation of fat, separation of the connective tissue fraction or without it; and isolation of the isolate (surimi) or the preparation of myofibrillar proteins with the tendinous fraction (Laage et al. 2010).
At present, MRPM is a relatively common component of cheap, finely comminuted thermally processed meat products of low shelf life. This is obviously connected with the economic aspect, i.e. the low price of such a raw material. In turn, the scope of applications for isolates of myofibrillar proteins (poultry surimi) is expanded to include the potential substitution of chicken breast muscle and to use it as a valuable binder in restructured products or as an addition to more expensive and less perishable meat products. Moreover, new high-protein products may be obtained using this preparation as the main protein matrix component. Other solutions include the application of the myofibril protein isolate from the meat of chickens, after their laying period as an addition to the injection of brine (Li 2006) or to extruded products combining plant and animal origin materials (Stangierski and Kijowski 2007).
An adequate technological quality of the raw myofibril preparation (MP) is very important, as it modifies the functional properties of thermally formed gels. Enzymatic methods are being increasingly used to control the technological properties of food raw materials (Rossi-Márquez et al. 2014; Ribota et al. 2010; Roccia et al. 2012). In the meat industry, proteins are cross-linked using transglutaminase (EC 2.3.2.13) (TG), both endogenous, found in fish meat, and supplemented as an additive in the course of processing. This makes it possible to produce novel meat products such as products from restructured meat (hamburgers, poultry meat balls, shao-mai), chicken kebab or various types of sausages (Tseng et al. 2000; Kilic 2003; Murphy et al. 2004; Serrano et al. 2004; Perlo et al. 2006). Muscle proteins, which as a result of thermal processing do not form a gel with the required rheological attributes, may be gelled using this enzyme. Enzymatic modification of proteins makes it possible to modify their technological quality to provide desirable, uniform and, what is even more important, repeatable functional properties.
The effect of transglutaminase on the final quality of the gel product has been thoroughly investigated. However, there is limited available information on the behaviour of water molecules in a modified protein system, subjected to heating and cooling, which is typical of the meat industry. Determination of the functional characteristics of enzymatically modified MP is only possible with the application of physical analyses sufficiently sensitive to follow changes in intramolecular interactions taking place in the investigated systems. They should supply information on molecular mechanisms in protein systems, while at the same time preserving the microstructure of the biopolymer system. An example of such a method is a technique used to identify interactions between water molecules in the tested system, i.e. nuclear magnetic resonance (NMR), particularly the so-called low-field relaxation. It results from a review of available literature that low-field NMR has an increasing number of applications in tested food materials due to the non-invasive character of the measurement (Andersen and Rinnan 2002; Bertram et al. 2002a; Pitombo and Lima 2003).
The intensity of the signal is proportional to the numbers of nuclei found in the system. The signal decay rate reflects molecular mobility in the investigated system (microdynamics) (Pedersen et al. 2001; Bertram et al. 2002b, 2004). The return process from the state of excitation to the state of equilibrium is referred to as spin–lattice relaxation, described by time T 1. In the case of the surimi-type protein preparation, we deal with a system, in which we observe the presence of large molecules. Water molecules interact with molecules of proteins by binding, and the process of water transfer in the system is slowed down. In turn, spin–spin relaxation time (T 2) reflects interactions of neighbouring spins, even within the same macromolecule. In the case of a protein preparation, i.e. a substance of high viscosity, field components induced by neighbouring nuclei are considerable and time T 2 is significantly shorter than time T 1 (Laurent et al. 2000).
This study is devoted to an analysis of the effect of transglutaminase on myofibril proteins in chicken muscles at the molecular level, which significantly affect the functional and technological quality of poultry surimi. For this reason, the aim of this study was to analyse changes in the degree of water binding by the protein system during heating and cooling of a poultry preparation subjected to the catalytic action of transglutaminase. The tested temperature ranges were typical of the conditions found in commercial scale meat processing.
Materials and Methods
Sample Preparations
The raw material for analyses comprised meat mechanically recovered from carcasses of broiler chickens, trimmed to remove breast and leg muscles and obtained using a Lima RM 500 apparatus (Lima S.A.S.; Quimper, France). The meat material was purchased at a local poultry abattoir and processing plant (Zakłady Drobiarskie “Koziegłowy” Sp. z o.o., Poland). The temperature of MRPM immediately after separation did not exceed +6 °C. After mixing the material to make it more homogenous, the prepared batch of raw material was divided into portions weighing about 1 kg, packed into polyester bags and frozen in industrial conditions to the temperature of −35 °C. Prior to experiments, samples were defrosted at the temperature of 4–5 °C for the period of approximately 18 h. The method of myofibril preparation (MP) production was adopted following the procedure described in a patent claim (Kijowski et al. 1996). Meat material was washed with a 1 % aqueous sodium chloride solution (0.169 mol/L) and then water (MRPM/water, 1:3 w/v), and fat and the connective tissue were separated on sieves. Obtained MP samples were modified using an addition of 3 g/kg of enzymatic preparation for 5 h at 6–7 °C. The amount of the applied transglutaminase supplementation as well as the treatment duration of the protein preparation was established on the basis of earlier research results (Stangierski and Kaczmarek 2012). For this purpose, a commercial preparation ACTIVA WM by Ajinomoto Co., Ltd. (Barentz, Poland) was used. The preparation contained 1 % microbial transglutaminase (MTG; Streptoverticillium sp.) with an activity of 100 U/g preparation and 99 % maltodextrin serving as the enzyme carrier (Ajinomoto’s specifications). One unit was the amount of the enzyme which catalysed the formation of 1 μmol of hydroxamic acid per minute at 37 °C.
Determination of Dynamics of Water Binding Using the 1H NMR
The poultry surimi samples were placed into 8-mm NMR tubes that were then sealed with Parafilm® to prevent moisture loss during the experiment. The NMR measurements were performed using a PS 15T pulse spectrometer operating at 15 MHz (Ellab, Poznań, Poland) and equipped with an integral temperature control system.
The inversion recovery pulse sequence (π − τ − π/2) was applied to the spin–lattice relaxation time T 1 experiment. The distance between the RF pulse (τ) ranged from 20 to 3500 ms; the repetition time TR was from 20 s. During each measurement of spin–lattice relaxation time, 32 signals of free induction delay (FID) and 110 points on each FID signal were collected. For the measurements of spin–spin T 2, the Carr–Purcell–Meiboom–Gill (CPMG) pulse train was used (Carr and Purcell 1954; Meiboom and Gill 1958). The distance between spin echoes was adjusted depending on the temperature. It changed from 2 ms (20 °C) to 8 ms (70 °C). The amplitudes of 100 spin echoes were recorded. Five signal accumulations were applied with a repetition time of 20 s. The rate of heating/cooling was 2 °C/min.
For temperature dependencies of relaxation times, the measurements were recorded at temperatures increasing from 20 to 70 °C and then at temperatures decreasing to 20 °C, at every 10 °C or 5 °C (for higher temperatures) after a 20-min stabilisation. The time dependencies of relaxation times were analysed every 24 h for 7 days after cooling at 20 °C. The parameters of pulse sequences (inversion recovery and CPMG) were the same as for the measurement of 20 °C. Between the experiments, the samples were stored at 5 °C. Both measurements of temperature and time dependencies of T 1 and T 2 were recorded for the same samples. This procedure provides an analysis without the need to take into account individual characteristics of the biological material.
All the determinations were performed in two replications. Experiments were carried out in two variations. This refers to the systems with and without the addition of transglutaminase. Each time, fresh protein preparations were prepared in accordance with the procedure described earlier. The data presented in the figures are the result of averaging the results obtained in the individual procedures. Mean values as well as standard deviations were calculated with the assistance of the STATISTICA PL v. 10.0 software by StatSoft.
Results and Discussion
Changes in T 1 and T 2 During Heating
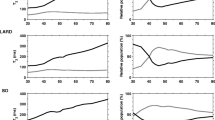
Values of spin–lattice relaxation times T 1 depend on the relative proportions between free and bound water in the tested biological system. The material containing more free water is characterised by a longer relaxation time. In the course of heating in the investigated protein systems, three characteristic ranges of temperatures were distinguished (Fig. 1a). Similar temperature dependencies of relaxation times were observed in the case of heating of starch–water systems (Baranowska et al. 2010, 2011). In the first area, relaxation time T 1 increased with an increase in heating temperature. The molecular dynamics of water increased as a result of the increase in temperature. For the control system (MP), the maximum T 1 was found at 50 °C. Reduction of the spin–lattice relaxation time at temperatures exceeding 50 °C, observed in the second area, should be attributed to the process of protein denaturation and the formation of the gel network. Temperature changes in proteins of various origins were described in more detail in earlier publications (Goetz and Koehler 2005; Stangierski and Baranowska 2008; Stangierski et al. 2012). On all thermograms generated by differential scanning calorimetry (DSC), two basic peaks of changes were recorded, where T1 (for myosin) corresponded to the temperature range from 56.94 °C to 57.79 °C. In the case of T2 (for actin) temperatures differences in the transition temperatures were for all incubation times, fell within the range from 70.18 °C to 70.81 °C. The presented values of temperatures refer to the MP with an addition of enzymatic preparation. The mean values of T1 and T2 were 57.65 °C and 70.62 °C, respectively for the control sample (Stangierski et al. 2013).
a Temperature changes in spin–lattice relaxation times during heating (from 20 to 70 °C) of myofibril preparation (MP) samples. The values are expressed as means ± SD (n = 6). b Temperature changes in spin–spin relaxation times during heating (from 20 to 70 °C) of myofibril preparation (MP) samples. The values are expressed as means ± SD (n = 6)
Conformation changes in proteins, manifested in the reduced values of T 1 with an increase in temperature, are observed at a temperature range of 50–60 °C. Above this temperature, in the third area, the systems are characterised by a repeated increase in T 1 values with an increase in heating temperature. The temperature of changes in the spin–lattice relaxation time in the first and third areas is caused solely by thermal proton movement. The second area, in which despite an increase in temperature the relaxation time is reduced, is connected with the process of water binding in the forming protein network (Tornberg 2005).
Probably due to the low ionic strength of the system, the molecules of myosin and actin are not dissolved but linked only through filament structures. Thus, in the analysed system, we may find both free molecules of myosin and actin as well as actin–myosin complexes. Cross-linking of proteins occurring at low temperatures probably results from interactions between heads of some free myosin molecules and myosin and actin. Wang and Smith (1994a) identified in the myosin isolated from the chicken breast muscle ten different protein domains. Smyth et al. (1996) showed that the domain containing α-helix of light meromyosin was unfolded as a result of heating within the temperature range of 20–46 °C. This suggests a thermal process of partial unfolding of myosin, as a result of which new, previously unavailable hydrophilic and hydrophobic groups were revealed. This led to an increased capacity of interactions between its chains, on the one hand via disulfide bonds and on the other hand via embedded water molecules and their attachment via hydrogen bonds with polar radicals of amino acids, which to a considerable degree are found on the surface of protein molecules (Niwa 1992). Intensification of structuring processes, resulting from the partial unfolding of myosin, is particularly evident in systems containing transglutaminase (Yongsawatdigul and Piyadhammaviboon 2005). As a result of the catalytic action of transglutaminase, cross linkages are formed between basic peptide groups of proteins, and they are bound with no involvement of water. Partial unfolding of the structure of muscle proteins, induced either enzymatically or thermally, results in the formation of a spatial protein network of varying concentrations of segments, exhibiting strong elastic properties.
Water molecules are actively involved in the formation of gel structure. The fraction of bound water has a markedly reduced mobility as a result of ionic or hydrogen bonding with polymer macromolecules (Laage et al. 2010). In turn, bulk water found in the system interacts with the fraction of bound water through a rapid chemical exchange of protons. This is indicated by one spin–lattice relaxation time. In comparison to the control system in enzymatically modified samples, the maximum T 1 was recorded at a lower temperature (Fig. 1a). The displacement of T 1 minimum was probably the result of protein denaturation changes of the examined system in the presence of microbial origin transglutaminase preparation (MTGP). This seems to be corroborated by the thermodynamic analysis (DSC) of the myofibril preparation modified by transglutaminase. In the case of samples with the addition of enzymatic preparation, we can see a certain decrease in the T1 (for myosin) temperature by approximately 1 °C (after 5-h modification) in relation to the control sample (Stangierski et al. 2013). The effect of the transglutaminase addition on thermal properties of proteins was similar to the results of investigations conducted by Aktaş and Kiliç (2005) on myofibril preparations obtained from beef. A much greater reduction in transformation temperatures of myofibril proteins obtained from bovine hearts under the influence of the enzyme was reported by Ramirez-Suarez et al. (2001). A transglutaminase addition resulted in a statistically significant reduction in the heat of transformation corresponding to myosin. A decrease in transformation temperature of myosin may indicate lower thermostability of the system.
It needs to be stated here that the addition of microbial origin transglutaminase preparation (MTGP) results in the shift of the range of temperatures connected with the denaturation processes and the formation of protein network towards temperatures lowered by several degrees. However, it is not a drastic reduction of these values. This was also observed in studies using differential scanning calorimetry (DSC) (Kazemi et al. 2011; Stangierski et al. 2013) and rheological analyses (Stangierski et al. 2014).
Measurements of spin–spin relaxation times confirm earlier observations concerning the process of protein gelation occurring in unheated MP and MP + MTGP systems (Stangierski and Baranowska 2008). The onset of structural changes in protein systems is manifested in the separation of spin–spin relaxation time into two components (Fig. 1b). Components of relaxation time are referred to as the short component (T 22), for lower values, and the long component (T 21), describing greater values. Components of relaxation time reflect molecular dynamics of bound water (T 22) and bulk water (T 21) (Chan-Ick et al. 2011). The short component, similarly to time T 2 at lower temperatures, is characterised by a slight reduction of values. The long component of spin–spin relaxation time shows a drastic increase in values from 55 °C for the control (MP) and 50 °C for the sample with the addition of MTGP. This is related to changes in the organisation of water in the protein system as a result of myosin denaturation, which probably initiates the formation of the gel network (Lee 1984). Similar results were recorded when observing a considerable increase in values of the modulus of elasticity G 1 at temperatures above 65 °C, occurring in both tested systems. It is connected with further structural changes in myosin molecules (Stangierski et al. 2014). At a temperature above 70 °C, significant changes also occur in the G-actin monomer (Ziegler and Acton 1984). An increase in elasticity of myosin gel within the range of 51–80 °C is caused first of all by the involvement of heads of myosin molecules. Within the range of 60–65 °C, the network structure is strengthened by a large number of actin–myosin complexes (Yasui and Samejima 1990). Probably, this effect is enhanced by the catalysing effect of transglutaminase. Sulfhydryl groups (−SH) oxidised during heating at a temperature above 70 °C lead to the formation of strong S–S covalent bonds in proteins. This rapid increase in the modulus of elasticity above 65 °C is also influenced by changes in structures observed within the hydrocolloid continuous phase as a result of thermal changes in sarcoplasmic proteins (Wright et al. 1977).
Changes in T 1 and T 2 Values During Cooling
Analyses conducted during the cooling process in both systems showed only a reduction of T 1 values with a decrease in temperature (Fig. 2a). Values of the short component T 22 of spin–spin relaxation time did not change significantly over the entire analysed range of temperatures. The small decrease of short components of T 22 to about 45 °C could have been connected with the temperature effects in dynamics of water molecules (Fig. 2b). It was also found that the system with transglutaminase is characterised by markedly greater values of the T 22 component.
a Temperature changes in spin–lattice relaxation times during cooling (from 70 to 20 °C) of myofibril preparation (MP) samples. The values are expressed as means ± SD (n = 6). b Temperature changes in spin–spin relaxation times during cooling (form 70 to 20 °C) of myofibril preparation (MP) samples. The values are expressed as means ± SD (n = 6)
The process of MP cooling showed a maximum at 55 °C for the preparation with transglutaminase (MP + MTGP). In turn, the long component of the spin–spin relaxation time in the MP system decays at low temperatures. Below 40 °C, the system has one spin–spin relaxation time. These differences probably need to be attributed to the different courses of protein gel stabilisation.
Earlier studies by Stangierski et al. (2014) showed that during sample cooling from the temperature of 76 to 23 °C, the modulus of elasticity G 1 in the control system increased monotonically. This may be connected with transformations within the intermolecular hydrogen bonds. Wang and Smith (1994b) showed that in myosin isolated from chicken breast muscles, heated to a temperature above 55 °C and next cooled to 25 °C, hydrogen bonds were formed in the β-structure of proteins. It contributes to the stabilisation and increased elasticity of the actin–myosin gel lattice and an enhancement of its elastic properties.
Cooling of the tested protein systems, containing transglutaminase to approximately 35 °C, leads to a slight, linear reduction of G 1 values (Stangierski et al. 2014). This may be connected with a weakening of hydrophobic interactions accelerating with a reduction of the temperature in the system. This is suggested by changes in elasticity, which are entropic in character as it is characteristic of covalently bound polymers (Case et al. 1992). Below 35 °C, similarly as was the case in the control system, the spatial gel network is stabilised by hydrogen bonds. This is reflected in the values of the lost tangent. In the MP system with an addition of MTGP, more water is found outside the lattice, in the hydrocolloid phase. This phase is not involved in the modification of the elastic property of the entire system but is rather a viscous agent.
The addition of MTGP promotes protein–protein interaction very effectively, particularly at higher temperatures. This is manifested in the observed increase in the mobility of molecules of water unbound with the protein matrix. Under identical temperatures, the absence of this enzyme results in the dominance of protein–water interactions over protein–protein interactions. Thus, the reduction of T 22 values observed for water molecules is connected with a limitation of freedom of mobility.
Determination of Activation Energy Values
The utilisation of low-field NMR to analyse the molecular dynamics of water in biological systems, apart from the quantitative (T 1) and qualitative (T 2) description, makes it also possible to determine the mean correlation time for rotational and translational movements of water molecules. It is a parameter which definitely characterises mobility at the molecular level. If relaxation times meet the condition, T 1 > T 2, then the correlation time varies within the range from 10−9 to 10−7 s (Coyle et al. 1996). Measured spin–lattice and spin–spin relaxation times made it possible to determine the mean correlation time τ c for the rotational movement of water molecules in the system (Tang and Belton 1999). Determination of τ c values for individual surimi systems was provided by the analytical solution of the equation describing frequency dependencies of relaxation times in terms of this parameter. In turn, temperature dependencies of the mean correlation times take the Arrhenius form:
where ΔE a is the activation energy of molecule reorientation, R is a gas constant, T is the absolute temperature, and τ 0 is the pre-exponential factor corresponding to the correlation time at an infinite temperature.
Values of activation energy ΔE a in the heating and cooling processes of the tested systems were determined based on experimental data and calculations. During the heating of protein preparations, a structural transition was observed. It was also observed that the onset of conformation changes in the system with the addition of transglutaminase was recorded at a temperature of approximately 45 °C, while in the control system, it was above 50 °C (Figs. 1a, b). Relaxation studies showed that above the transition temperature, two proton fractions were observed, relaxing at different spin–spin relaxation times. Based on temperature dependencies of mean correlation times, the values of activation energy were calculated for the water molecule reorientation in the system. Below the transition temperature, this energy amounts to 5.1 kJ/mol for the protein preparation and 4.1 kJ/mol for the preparation with an addition of the enzyme. For the proton fraction, revealed as a result of structural changes, a considerable increase was observed for values of activation energy. In the protein preparation, this energy was 16.8 kJ/mol, while in the system of proteins modified enzymatically, it was 18.3 kJ/mol. At low temperatures, the small and only slightly different values of activation energy indicate that reorientation of water molecules is inhibited in neither of these two systems. The addition of the enzyme reduces the value the of energy barrier. This is probably related, among other things, with the effect of the displacement of water from the protein system as a result of the presence of transglutaminase. This negative effect of transglutaminase on animal protein systems is probably a consequence of the reduced capacity of water binding by proteins soluble in salt, resulting from the formed cross-linkages. Such an effect was observed at an excessive addition of the enzyme and at a too high temperature of the cross-linking reaction (40 °C) (Ruiz-Carrascal and Regenstein 2002; Stangierski and Kaczmarek 2012). Values of activation energy for poultry protein systems determined at high temperatures were comparable to those recorded for starch systems (Baranowska et al. 2010). This indicates the formation of the polymer network, in which mobility of water molecules is strongly inhibited as a result of the binding of some water molecules on the surface of proteins.
In the course of system cooling, at a temperature above 50 °C, the activation energy of free water in the protein system was 7.6 kJ/mol and it was approximately two times smaller than during heating. In the area of low temperatures (below 50 °C), where the relaxation time component describing the molecular dynamics of bulk water declines, activation energy decreases to 3.6 kJ/mol. Such a low value of this parameter indicates that heating and cooling of the tested protein preparation result in the evacuation of water outside the network of denatured protein. Cooling of the system with the enzyme is also reflected in the lower value of activation energy at high temperatures (13.6 kJ/mol). However, it is a markedly higher value than that recorded for the control. This may suggest a significant effect of the enzyme on water binding processes also in the course of sample cooling. In this system, we observe the presence of two independent water fractions over the entire range of temperatures (Fig. 2b). At a temperature below 45 °C, values of activation energy for molecular movement in both fractions are similar—4.7 kJ/mol for bound water and 3.0 kJ/mol for bulk water.
Both relaxation times were measured jointly within 168 h (7 days) after both systems had cooled. The beginning of the measurements (t = 0) was assumed to be the moment at which the system reached +20 °C after cooling. In the MP system with the addition of the enzyme over the entire analysed period, two components of spin–spin relaxation time were observed (Fig. 3a). This effect was not recorded in the control system. This may mean that the addition of MTGP caused a change in the organisation of water in the system. The short component of T 2 in the PM + MTGP system has slightly lower values than relaxation time T 2 of the control. Thus, in the PM + MTGP system, one of the water fractions is slightly less mobile than water in the control system. Lower mobility is probably connected with a lower number of water molecules bound with proteins. The other water fraction, i.e. bulk water, is found in the system and exhibits considerable mobility. These results seem to confirm the theory that transglutaminase, under certain conditions, is a factor causing displacement of water from the system, rather than its binding. This is also confirmed by the results of temporal changes in T 1 values (Fig. 3b). We may observe a considerable increase in values of this parameter in comparison to the control system measured over the entire period of the analyses. A threefold increase of the value T 21 and about 15 % increase of the value T 1 imply that in time to 72 h, a myofibril preparation with transglutaminase increases the quantity of bulk water. This can be explained as a change in protein system. An increase in the distance between thin and thick filaments was reflected in the results of the analyses conducted using NMR as an increase in relaxation times (Bertram et al. 2004). Relaxation times recorded in another study by Bertram et al. (2006) indicate that the main changes in the characteristics of water contained in porcine meat tissue, observed during heating within the range of 25 °C up to 75 °C, take place between 40 and 50 °C. At the same time, the authors of that study indicated a correlation between denaturation of myosin tails and light chains at approximately 53–58 °C and thermally induced changes in the degradation of water found in myofibrils, based on DSC and NMR analyses.
An important technological conclusion may be derived from the analysis of spin–lattice relaxation times and the long component of the spin–spin relaxation times conducted for the enzymatically modified MP sample. The addition of transglutaminase to proteins leads to the binding of water in the system for the first 48 h after cooling. Next, we observe a considerable increase in T 1 values. Maxima of the above mentioned relaxation times in the MP system modified with transglutaminase were observed 72 h after cooling. After that time, the values of both parameters decrease, reaching the values recorded 24 h after cooling. Probably, in the system, a temporal change in the dynamic status of water molecules is connected with the long-term action of transglutaminase. The initial elongation of relaxation times indicates a displacement of water from the system. This means that in the case of these cold-stored samples after 3 days, we may expect a significant drip of water from these samples. The control MP is characterised by a reduction of T 1 values up to 96 h after cooling. This shows that in the analysed period, water binding takes place in the system. After that time, a slight increase is observed in T 1. Similarly to the MP + MTGP system, after 7 days, the T 1 values are the same as at 24 h after cooling. Values of spin–spin relaxation times are constant. Additionally, the system non-modified enzymatically at 24 h after cooling has one spin–spin relaxation time.
Conclusion
The adopted analytical technique was to provide insight into molecular mechanisms accompanying the formation of spatial protein structures and their changes under the influence of thermal processing. Aiming at the determination of the essence of molecular transformations accompanying the formation of spatial structure protein, we analysed the kinetics of structuration processes. The molecular mechanism responsible for the phenomena described above is complex. This results from the fact that the protein preparation produced from MRPM in reality, apart from the actin–myosin complex and other myofibrillar proteins, also contains certain amounts of sarcoplasmic proteins and connective tissue. Thermal processing contributes to the formation of a new physical system of proteins, characterised by the complete change of the initial structural parameters of all its components. The presence of transglutaminase in the protein system results in a situation when the process of changes begins and ends at lower temperatures. Denaturation of proteins in the presence of the enzyme is manifested in an increase of activation energy values. This is connected with the displacement of a portion of the water molecules outside the formed biopolymer network. This results in the increase of spin–lattice relaxation times observed during sample cooling in the system of the enzymatically modified myofibril preparation. This is also confirmed by the twofold lower value of activation energy for the molecular movements of protons. The conducted analyses of heated and then cooled samples clearly showed that the addition of MTGP to the MP system causes displacement of water outside the protein network formed during gelation. Analysis of values of relaxation time T 1 showed that the addition of the enzyme to MP causes water binding in the sample up to 48 h after gel cooling. This means that after 3 days of cold storage, a significant water drip may take place from modified protein gels. The effect of the enzyme, observed earlier at the macroscopic level, provided grounds only for the statement on an increase in network rigidity. Analyses at the submolecular level showed that the mechanism of water displacement observed, e.g. after several days of storage of gels, produced from enzymatically modified MP is probably caused by the competitiveness of protein–protein and protein–water bonds in the polymer network.
References
Aktaş, N., & Kiliç, B. (2005). Effect of microbial transglutaminase on thermal and electrophoretic properties of ground beef. LWT--Food Science and Technology, 38, 815–819.
Andersen, C. M., & Rinnan, A. (2002). Distribution of water in fresh cod. LWT--Food Science and Technology, 35, 687–696.
Baranowska, H. M., Sikora, M., Krystjan, M., & Tomasik, P. (2010). Contribution to understanding gelatization of granular starch. In M. Fiedorowicz & E. Bertoff (Eds.), Recent advances in biopolymer science and technology (ch.1, pp 13–28). Cracow: Polish Society of Food Technologist.
Baranowska, H. M., Sikora, M., Krystyjan, M., & Tomasik, P. (2011). Analysis of the formation of starch—hydrocolloid binary gels and their structure based on the relaxation times of the water molecules. Polimery, 56, 478–483.
Bertram, H. C., Donstrup, S., Karlson, A. H., & Andersen, H. J. (2002a). Continuous distribution analysis of T2 relaxation in meat—an approach in the determination of water-holding capacity. Meat Science, 60, 279–285.
Bertram, H. C., Rasmussen, M., Busk, H., & Oksbjorg, N. (2002b). Changes in porcine muscle water characteristics during growth—an low-field NMR relaxation study. Journal of Magnetic Resonance, 157, 267–276.
Bertram, H. C., Engelsen, S. B., Busk, H., Karlson, A. H., & Andersen, H. J. (2004). Water properties during cooking of pork studied by low-field NMR relaxation: effects of curing and the RN− gene. Meat Science, 66, 437–446.
Bertram, H. C., Wu, Z., Berg, F., & Andersen, H. J. (2006). NMR relaxometry and differential scanning calorimetry during meat cooking. Meat Science, 74, 684–689.
Carr, H. Y., & Purcell, E. M. (1954). Effects of diffusion on free precession in nuclear magnetic resonance experiments. Physics Review, 94, 630–638.
Case, S. E., Knopp, J. A., Hamann, D. D., & Schwartz, S. J. (1992). Characterization of gelation of konjac manna using lyotropic salts and rheological measurements. In P. A. Williams & D. J. Wedlock (Eds.), Gums and Stabilizers for the Food Industry (6th ed., pp. 489–500). New York: Oxford University Press.
Chan-Ick, C., Hye-Won, W., & Myong-Soo, C. (2011). Caking characteristics and sensory attributes of ramen soup powder evaluated using a low-resolution proton NMR technique. Food Research International, 44, 1102–1107.
Cortez-Vega, W. R., Fonseca, G. G., & Prentice, C. (2012). Comparisons of the properties of Whitemouth Croaker (Micropogonias furnieri) surimi and mechanically deboned chicken meat surimi-like material. Food and Nutrition Sciences, 3, 1480–1483.
Cortez-Vega, W. R., Fonseca, G. G., & Prentice, C. (2015). Optimization of parameters for obtaining surimi-like material from mechanically separated chicken meat using response surface methodology. Journal of Food Science and Technology. doi:10.1007/s13197-013-1056-1.
Coyle, F. M., Martin, S. J., & McBrierty, V. J. (1996). Dynamics of water molecules in polymers. Journal of Molecular Liquids, 69, 95–116.
EFSA Panel on Biological Hazards (BIOHAZ). (2013). Scientific Opinion on the public health risks related to mechanically separated meat (MSM) derived from poultry and swine. EFSA Journal, 11(3), 3137. p. 78.
Goetz, J., & Koehler, P. (2005). Study of the thermal denaturation of selected proteins of whey and egg by low resolution NMR. LWT--Food Science and Technology, 38, 501–512.
Kazemi, S., Ngadi, M. O., & Gariépy, C. (2011). Protein denaturation in pork Longissimus muscle of different quality groups. Food and Bioprocess Technology, 4, 102–106.
Kijowski, J., Stangierski, J., Magnuski, T., & Pikul, J. (1996). Method of obtaining myofibril concentrate. Polish Patent Pl, 169727, B1.
Kilic, B. (2003). Effect of microbial transglutaminase and sodium caseinate on quality of chicken doner kebab. Meat Science, 63, 417–421.
Laage, D., Stimemann, G., & Hynes, J. T. (2010). Water reorientation in the hydration shells of hydrophilic and hydrophobic solutes. Science China Physics Mechanics and Astronomy, 53, 1068–1072.
Laurent, W., Bonny, J. M., & Renou, J. P. (2000). Muscle characterization by NMR imaging and spectroscopie techniques. Food Chemistry, 69, 419–426.
Lee, C. M. (1984). Surimi process technology. Food Technology, 38(11), 69–74.
Li, C. T. (2006). Myofibrillar protein extracts from spent hen meat to improve whole muscle processed meats. Meat Science, 72, 581–583.
Meiboom, S., & Gill, D. (1958). Modified spin-echo method for measuring nuclear relaxation times. Review of Scientific Instruments, 29, 688–691.
Murphy, S. C., Gilroy, D., Kerry, J. F., Buckley, D. J., & Kerry, J. P. (2004). Evaluation of surimi, fat and water content in a low/no added pork sausage formulation using response surface methodology. Meat Science, 66, 689–701.
Negrão, C. C., Mizubuti, I. Y., & Morita, M. C. (2005). Biological evaluation of mechanically deboned chicken meat protein quality. Food Chemistry, 90, 579–583.
Niwa, E. (1992). Chemistry of surimi gelation. In T. C. Lanier & C. M. Lee (Eds.), Surimi technology (pp. 389–427). New York: Marcel Dekker, Inc.
Nowsad, A., Kanoh, S., & Niwa, E. (2000). Thermal gelation characteristics of breast and thigh muscles of spent hen and broiler and their surimi. Meat Science, 54, 169–175.
Pedersen, H. T., Lundby, B. F., & Engelsen, S. B. (2001). The multivariate advantage in fat determination in meat by bench-top NMR. Innovative Food Science and Emerging Technologies, 2, 87–94.
Perlo, F., Bonato, P., Teira, G., Fabre, R., & Kueider, S. (2006). Physicochemical and sensory properties of chicken nuggets with washed mechanically deboned chicken meat: research note. Meat Science, 7, 785–788.
Pitombo, R. N. M., & Lima, G. A. M. R. (2003). Nuclear magnetic resonance and water activity in measuring the water mobility in pintado (Pseudoplatystoma) fish. Journal of Food Engineering, 5, 59–66.
Ramadhan, K., Huda, N., & Ahmad, R. (2011). Effect of number and washing solutions on functional properties of surimi-like material from duck meat. Journal of Food Science and Technology. doi:10.1007/s13197-011-0510-1.
Ramirez-Suarez, J. C., Xiong, Y. L., & Wang, B. (2001). Transglutaminase cross-linking of bovine cardiac myofibrillar proteins and its effect on protein gelation. Journal of Muscle Foods, 12, 85–96.
Rbotta, P. D., Pérez, G. T., Añón, M. C., & León, A. E. (2010). Optimization of additive combination for improved soy–wheat bread quality. Food and Bioprocess Technology, 3, 395–405.
Roccia, P., Ribotta, P. D., Ferrero, C., Pérez, G. T., & León, A. E. (2012). Enzymes action on wheat–soy dough properties and bread quality. Food and Bioprocess Technology, 5, 1255–1264.
Rossi-Márquez, G., Di Pierro, P., Esposito, M., Mariniello, L., & Porta, R. (2014). Application of transglutaminase-crosslinked whey protein/pectin films as water barrier coatings in fried and baked foods. Food and Bioprocess Technology, 7, 447–455.
Ruiz-Carrascal, J., & Regenstein, J. (2002). Emulsion stability and water uptake ability of chicken breast muscle proteins as affected by microbial transglutaminase. Journal of Food Science, 67(2), 734–739.
Serdaroğlu, M., & Turp, G. Y. (2005). Effects of deboning methods on chemical composition and some properties of beef and turkey meat. Turkish Journal of Veterinary and Animal Sciences, 29, 797–802.
Serrano, A., Cofrades, S., & Colmenero, F. J. (2004). Transglutaminase as binding agent in fresh restructured beef steak with added walnuts. Food Chemistry, 85, 423–429.
Smyth, A. B., Smith, D. M., Vega-Warner, V., & O'neill, E. (1996). Thermal denaturation and aggregation of chicken breast muscle myosin and subfragments. Journal of Agricultural and Food Chemistry, 44, 1005–1010.
Stangierski, J., & Baranowska, H. (2008). Analysis of texture and dynamics of water binging in enzymatically modified myofibrillar preparation obtained from washed mechanically recovered poultry meat. European Food Research and Technology, 226, 857–860.
Stangierski, J., & Kaczmarek, A. (2012). Effects of transglutaminase modification on the quality of poultry surimi. Investigations on poultry surimi obtained from mechanically recovered chicken meat. Fleischwirtschaft International, 6, 69–73.
Stangierski, J., & Kijowski, J. (2000). Optimization of conditions for myofibril preparation from mechanically recovered chicken meat. Nahrung/Food, 44(5), 333–338.
Stangierski J., & Kijowski J. (2007). The application of dried myofibril preparation from poultry meat in the production of corn puffs. XVIII European Symposium on the Quality of Poultry Meat. Czech Republic, Prague 2–5 September, 325–326.
Stangierski, J., Rezler, R., Baranowska, H. M., & Poliszko, S. (2012). Effect of enzymatic modification on chicken surimi. Czech Journal of Food and Sciences, 30(5), 404–411.
Stangierski, J., Zabielski, J., & Grześ, B. (2013). Modification of functional quality of raw myofibril preparation obtained from water-washed mechanically recovered chicken meat. European Food Research and Technology, 236, 449–458.
Stangierski, J., Rezler, R., & Leśnierowski, G. (2014). Analysis of the effect of heating on rheological attributes of washed mechanically recovered chicken meat modified with transglutaminase. Journal of Food Engineering, 141, 13–19.
Tang, H., & Belton, P. S. (1999). Proton relaxation in plant cell walls and model systems. In P. S. Belton, B. P. Hills, & G. A. Webb (Eds.), Webb advances in magnetic resonance in food science (pp. 166–184). Cambridge, UK: Royal Society of Chemistry.
Tornberg, E. (2005). Effects of head on meat proteins—implications on structure and quality of meat products. Meat Science, 70, 493–508.
Trindade, M. A., Felicio, P. E., & Castillo, C. J. C. (2004). Mechanically separated meat of broiler breeder and white layer spent hens. Scientia Agricola, 61(2), 234–239.
Tseng, T. F., Liu, D. C., & Chen, M. T. (2000). Evaluation of transglutaminase on the quality of low-salt chicken meat balls. Meat Science, 55, 427–431.
Wang, S. F., & Smith, D. M. (1994a). Heat-induced denaturation and properties of chicken breast myosin and F-actin in the presence and absence of pyrophosphate. Journal of Agricultural and Food Chemistry, 42, 2665–2670.
Wang, S. F., & Smith, D. M. (1994b). Poultry muscle proteins and heat-induced gelation. Poultry Science Reviews, 5, 145–167.
Wright, D. J., Leach, I. B., & Wilding, P. (1977). Differential scanning calorimetric studies of muscle and its constituent proteins. Journal of Science and Food Agriculture, 28, 557–560.
Yang, T., & Froning, G. (1994). Evaluation of protein functionality in alkali and nonalkali surimi processed mechanically deboned chicken meat. Journal of Muscle Foods, 5, 221–232.
Yasui, T., & Samejima, K. (1990). Recent advances in meat science in Japan: functionality of muscle in gelation mechanism of structured meat products. Japan Agricultural Research Quaterly, 24, 131–237.
Yongsawatdigul, J., & Piyadhammaviboon, P. (2005). Effect of microbial transglutaminase on autolysis and gelation of lizardfish surimi. Journal of the Science of Food Agriculture, 85, 1453–1460.
Ziegler, G. P., & Acton, J. C. (1984). Mechanisms of gel formation by proteins of muscle tissue. Food Technology, 38(5), 77–82.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Stangierski, J., Baranowska, H.M. The Influence of Heating and Cooling Process on the Water Binding in Transglutaminase-Modified Chicken Protein Preparation, Assessed Using Low-Field NMR. Food Bioprocess Technol 8, 2359–2367 (2015). https://doi.org/10.1007/s11947-015-1618-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11947-015-1618-5