Abstract
Purpose of review
Cirrhosis is one of the most important global public health problems. Patients with cirrhosis risk progression to acute-on-chronic liver failure (ACLF), associated with high mortality rates, and development of hepatocellular carcinoma (HCC). Metabolomics could identify urgently required novel biomarkers to improve disease diagnosis, monitor progression, and identify therapies.
Recent findings
In this review, current metabolic studies in decompensated cirrhosis, ACLF, and HCC over the past 3 years are summarised. Over numerous metabolomics studies, in cirrhosis, common alterations in proteins, carbohydrates, lipids, bile acids, and microbial metabolites were identified. In ACLF, changes in metabolites related to energy metabolism, amino acids, lipids, bile acids, and microbial metabolites were reported. Amino acids, bile acids, free fatty acids, and phospholipids were identified as important metabolite classes for discrimination between cirrhosis and HCC.
Summary
Metabolomics can improve our understanding of advanced liver diseases and provide the basis of the future studies and therapeutic advancements.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cirrhosis causes more than 1.32 million deaths worldwide each year and is growing in incidence [1]. Cirrhosis is a frequent consequence of chronic liver injury related to different causes such as chronic viral hepatitis, chronic alcohol consumption (50%), and non-alcoholic fatty liver disease (NAFLD) (24%) [2,3,4]. The natural course of cirrhosis is characterised by an asymptomatic compensated stage followed by acute decompensation (AD) of the liver, which is indicated by the presence of variceal bleeding, ascites, hepatorenal syndrome, jaundice, or encephalopathy. Once patients develop acute decompensation, they require hospitalisation and have reduced 6-month survival. Three clinical courses were recently identified in patients with AD [5]. Patients with AD can develop unstable AD which often require hospitalisation. Other groups can have a more stable condition and infrequently require hospitalisation. In addition, the condition of patients with AD can subsequently deteriorate and progress to acute on chronic liver failure (ACLF). ACLF is characterised by acute deterioration of liver function and the development of multiple organ failure and high short-term morality [6, 7]. ACLF has poor prognosis which parallels acute liver failure (ALF), and patients can develop complications such as hepatic encephalopathy, coagulopathy, sepsis, and multiple organ failure [8]. Cirrhosis is also a primary risk factor for the development of hepatocellular carcinoma (HCC). The incident for development of HCC is higher in patients with cirrhosis compared to non-cirrhosis patients [9]. Therefore, early diagnosis, prognosis, and monitoring are important to prevent the progression of cirrhosis to life-threatening complications.
Metabolomics
Metabolomics is field of research which applies analytical chemistry techniques to the detection and measurement of low molecular weight metabolites (typically <1KDa) within a biological sample. Metabolic profiles are generated using advanced analytical platforms and can be used to gain insight of alterations into a specific condition or disease [10]. Compared to other omics, such as genomics and proteomics, metabolomics has the advantage of being a close reflection of molecular phenotype and reflects rapid changes associated with both acute and chronic pathophysiological conditions [11]. Therefore, metabolomics can detect alterations related to cells, tissues, or biochemical pathways associated with specific pathological diseases. Given the multiple metabolic pathways and processes governed in the liver, acute deteriorations in advanced liver disease may be reflected in abnormal metabolic profiles. This has the potential to be beneficial in clinical setting where it can be used to detect biomarkers for early diagnosis and monitoring of disease progression, thus providing insight about disease pathology [12].
Metabolomics can be applied on different types of samples such as tissues, cells, and biofluids [13]. Cell cultures can be utilised in metabolomics to understand cellular process whether through NMR or MS on cell extracts or supernatants. Tissue obtained from a specific organ can provide an indication of the physiological or pathological processes within a specific organ [14] as long as steps are undertaken to arrest metabolism at the point of sampling. Specialised biofluids such as saliva [15], exhaled breath condensate [16], and cerebrospinal fluid (CNS) [17] can also be utilised to understand metabolic changes within a specific organ. In clinical studies, most commonly used biofluids are urine and serum or plasma because they are less invasive to obtain and easy to prepare [18].
Metabolites are any small molecule (typically <1KDa) which can be produced by endogenous anabolic or catabolic processes such as amino acids, lipids, or organic acid. In addition, metabolites can be produced from exogenous sources such as food, drug, or microbial by-product.
Metabolic profiling can be divided into untargeted and targeted analysis. Untargeted analysis involves the comprehensive characterisation of as many metabolites within a biological sample as the technique allows and is usually used for new biomarker discovery [19]. Methods are usually generic and unbiased in terms of sample preparation and analytical conditions. Targeted analysis focuses on the detection of specific metabolites within biological samples, providing a more quantitative tool that can be developed for clinical use. Sample handling is designed to maximise recovery of the analytes of interest, and analytical conditions are optimised for these compounds. This provides a very sensitive and quantitative assay, which is often higher throughput than discovery approaches and requires less sample. Each approach has its advantages and disadvantages; more in-depth descriptions of these can be found in Cui et al. (2018) [20] and Griffiths et al. (2010) [21].
Metabolomics platforms
Different analytical platforms can be used to profile metabolites in biological samples, including nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry (MS), the latter coupled to either liquid (LC-MS) or gas chromatography (GC-MS). NMR is a non-destructive analytical platform, offering high reproducibility and requiring less sample preparation than MS techniques. MS provides both selectivity and sensitivity, resulting in improved metabolite discrimination and coverage [22]. When coupled with a separation technique, metabolites in a complex matrix can be separated before detection, improving detection and sensitivity further. LC-MS is more widely used in clinical studies than GC-MS because of the non-volatile nature of the sample required. Importantly, complete detection, identification, and quantification of metabolites cannot be achieved by a single platform. Each analytical platform has their own advantages, and the specific choice depends on many factors, including the sensitivity, selectivity, and reliability of the analytical platform which have been discussed in other reviews [23,24,25,26,27].
Data handling
The data generated by metabolic profiling contain large and complex information which—after pre-processing—can be analysed using different statistical and pattern recognition methods [28]. Typically, there are hundreds or even thousands of potential metabolites (variables) detected in a single sample. Univariate analysis (UVA) is applied to examine a single variable of interest across samples or sample classes, while multivariate analysis (MVA) can assess the relationship between multiple variables of interest. Commonly applied MVA include unsupervised analysis, such as principal components analysis (PCA), and supervised analysis such as partial least-squares discriminant analysis (PLS-DA) [28, 29]. These approaches can aid in the visualisation of class differences, outliers, and trends over time. Once important metabolites, such as amino acids or lipids, which discriminate between healthy and disease groups, have been identified, a biomarker predictive model can be generated. Models can be validated within the MVA software. Then, performance evaluation can be achieved with receiver operating characteristics (ROC) curve analysis [30]. The area under the curve (AUC) of the ROC can enable the identification of sensitive and specific biomarkers. Further studies will then need to be undertaken to validate these potential biomarkers in a new cohort.
The aim of this review is to summarise and assess recent metabolomic studies applied to advanced liver disease. The review will cover metabolomic studies in cirrhosis, ACLF, and HCC published within the past 3 years providing an overview of the potential metabolites which can be used for diagnosis, prognostication, and therapy.
Cirrhosis: potential biomarkers
Diagnostic markers of cirrhosis and progressive disease
Once patients develop cirrhosis, they have a high risk of progression to acute decompensation, which usually necessitates hospital admission. Therefore, it is important to identify early those patients who are at risk of developing cirrhosis or progression to acute decompensation or beyond to ACLF (Table 1). Yoo et al. (2019) [31] used untargeted LC-MS to identify potential biomarkers for classifying cirrhosis in serum samples obtained from patient with cirrhosis (n=94) and age- and sex-matched healthy controls (n=180). In this study, at baseline all subjects had normal liver functions and were monitored for 7 years. Those who developed cirrhosis were included in the cirrhosis group and those who did not develop cirrhosis were included in the healthy group. Therefore, any identified metabolic alterations in patients with normal function at baseline can potentially aid in understanding the mechanism of cirrhosis development. Important metabolic pathways which were significantly different between the two groups were related to glyoxylate and dicarboxylate metabolism, amino acid metabolism, fatty acid metabolism, linoleic acid metabolism, α-linolenic acid metabolism, and arachidonic acid metabolism.
Alterations of bile acids, amino acids, and free fatty acids have been shown to be associated with chronic liver disease regardless of the underlying cause [42, 43]. Identification of more specific markers can be more useful for the diagnosis and prognosis of liver cirrhosis. A 1H NMR study was conducted in serum of heathy control (n=25), HBV patients (n=20), and HBV patients with cirrhosis (n=25) to identify potential biomarkers to monitor the progression of cirrhosis in HBV patients [32]. Selected metabolites were considered good biomarkers when they showed AUC> 0.8. They suggest that different metabolites were associated with different stages of the disease. A potential panel of metabolites which was found to be associated with HBV progression to develop cirrhosis included acetate, formate, pyruvate, and glutamine in the serum. In a targeted LC-MS study, analysis was performed to predict stages of liver cirrhosis in serum from patients with HBV-associated liver cirrhosis (n=504) and healthy control (n=502) [33•]. Predictive models were constructed to diagnose patients with chronic liver disease from healthy controls, to differentiate fibrosis and cirrhosis and patients with advanced stage from early-stage cirrhosis. Then, a panel of metabolites was selected based on predictive models including taurocholate, tyrosine, valine, and linoelaidic acid. These metabolites were further validated in a second independent cohort showing AUC> 0.8 which confirmed the strength of their predictive models.
To distinguish patients with advanced stage cirrhosis, GC-MS and LC-MS were performed using plasma from HIV/HCV-coinfected (n=62) and HCV-monoinfected patients (n=28) [34]. No healthy control group was included in this study. The results showed increased fatty acids, bile acids, aromatic and sulphur amino acids, butyrate derivatives, oxidised phospholipids, energy-related metabolites, and bacterial fermentation-related metabolites in patients with advanced cirrhosis. In addition, they reported a decrease in lysophosphatidylcholines (LPC) and lysophosphatidylethanolamines (LPE), branched-chain amino acids (BCAA) and metabolites of tricarboxylic acid cycle in advanced stage cirrhosis. Glycolic acid, LPC (16:0), and taurocholic were selected as most significant metabolites for discriminating patients according to decompensation stage. AUC was used to evaluate the diagnostic performance which showed higher value of subsets of metabolites than for each metabolite separately [34]. This suggests that this panel of metabolites has the potential to be used as biomarkers for presence of acute decompensated cirrhosis. Another study showed increased concentrations of taurochenodeoxycholate, taurocholate, glycocholate, tyrosine, and PC (16:0/16:0) phosphatidylcholine in patients with advanced cirrhosis [35].
The contribution of bile acids to cirrhosis was assessed by targeted LC-MS, where 12 bile acids were measured in serum samples from healthy control (n=27) and cirrhosis (n=32) [36•]. They identified taurocholate (TCA) as the highest bile acid in patient with cirrhosis. The specific diagnostic and the therapeutic potential of TCA was investigated further in an in vitro study to find out whether this bile acid is a contributing factor to cirrhosis progression. They suggested that increased TCA in cirrhosis is mainly due to increased bile acid synthesis due to cholestasis. In addition, the mechanism of TCA contribution to cirrhosis was proposed to be through the effect of stellate cell activation via the TLR4 pathway.
Dysbiosis of the intestinal microbiome may also have an important role in advanced liver disease as a result of multiple interactions with the host immune system and gut epithelium. Alteration of gut microbiota and their metabolites have been investigated in liver cirrhosis because of their contribution to complications associated with cirrhosis, such as infection and hepatic encephalopathy [44, 45]. LC-MS was performed in urine from healthy control (n=47), compensated (n=49) and decompensated cirrhosis (n=46) to investigate the relationship between the gut microbiome and cirrhosis progression. Urine contains both host and microbial metabolites. Therefore, it can offer limited information about the interaction between host and microbial metabolites. The authors identified decreased level of microbiota-related metabolites, including N6-methyladnosine, 1-methyluric acid cinnamic acid, deconoylcarnitine, and phenacetylglutamine in urine which confirmed their finding of decreased microbiota responsible for sugar fermentation [37]. Adams et al. (2019) examined the link between bile acid profiles, gut microbiome, and cirrhosis in patients with NAFLD. They showed an association between alteration in bile acid profiles and changes in the composition of specific microbiota taxa in advanced cirrhosis [38].
Patients with cirrhosis are at increased risk for the development of complications such as hepatic encephalopathy (HE), hospitalisation, and death. Isolated hepatic encephalopathy is a common complication which can be observed in liver cirrhosis. In patients with cirrhosis, metabolic profiling was performed using urine to classify patient with cirrhosis who developed HE. Urinary metabolic profiling was found to be able to discriminate between patient with cirrhosis from healthy controls. Compared to healthy controls, patients with cirrhosis showed higher 1-methyl nicotinamide and lower hippurate, acetate, phenylacetylglycine, and N-methyl nicotinic acids. In addition, it was suggested that urinary metabolic profiling may have the potential to distinguish patients with overt hepatic encephalopathy from patients with no hepatic encephalopathy. However, in this study the models lacked validity due to the small sample size involved in this study [39].
Predicting poor outcome
In an exploratory study, GC-MS analysis was performed on serum and urine to identify potential metabolites which can predict outcomes in patient with cirrhosis such as hepatic encephalopathy (HE), transplant, hospitalisation, and death which they followed for 90 days [41]. Several metabolites have been suggested to discriminate between patients with and without outcome including saturated fatty acids, amino acids, and bioenergetic-related metabolites. Similar changes were found in serum and urine regardless of the specific complication.
Hepatorenal dysfunction is a serious complication which develops in patients with liver cirrhosis and is notable for the poor prognosis associated with this extra hepatic organ failure. Mindikoglu et al. (2018) conducted LC-MS on plasma from patients with cirrhosis (n=103) focusing on patients with hepatorenal dysfunction to predict mortality. They identified 34 metabolites which significantly predicted mortality in patients with cirrhosis including S-adenosylhomocysteine, glucuronate, trans-aconitate, 3-ureidopropionate, 3-(4-hydroxyphenyl)lactate, 3-methoxytyramine sulphate, arabitol/xylitol, N-formylmethionine, phenyllactate, and 7-methylguanine [40]. The identified metabolites were found to be independent predictors of glomerular filtration rate in the patient with cirrhosis. These identified metabolites should be taken forward to provide mechanistic understanding of potential metabolic mediators for the progression to hepatorenal dysfunction in liver cirrhosis.
ACLF: potential biomarkers
Patients with cirrhosis have a high risk for the development of ACLF. ACLF has short-term mortality estimated between 38 and 50% [46]. A challenge in clinical settings is to find a rapid method for early diagnosis, disease progression, and monitoring therapeutic outcome. Since ACLF can occur in all aetiologies, but with a diverse cause of deterioration, it is important to appraise these studies from this standpoint. A large number of clinical studies have focused on finding potential significant biomarkers to predict the development of ACLF and to understand the associated pathophysiology which can decrease the morbidity and mortality associated with ACLF (Table 2).
In a prospective multicentre study, untargeted LC-MS analysis was conducted on serum from patients with cirrhosis at the time of admission to identify possible metabolites for predicting ACLF development. They reported an increase in microbially derived metabolites and hormonal-related metabolites and a decrease in the level of phospholipids. These reported metabolites were found to be associated with the development ACLF and high mortality rate. In addition, they suggested that a combination of metabolites and relevant clinical parameters yields better prognostic biomarkers [47]. In another multicentre study, untargeted metabolomic was performed using LC-MS on serum samples to characterise the metabolomic profile associated with ACLF. Significant alterations in metabolic profile were found in metabolites associated with bioenergetic failure in ACLF, such as pentose phosphates. In addition, the identified metabolites correlated with concentrations of inflammatory markers, e.g. tumour necrosis factor α (TNF-α). They suggested that these changes were as result of the overwhelming inflammation in ACLF, mitochondrial dysfunction develop and led to development of organ failure [48•].
Lipid mediators can have regulatory effects on the systemic inflammatory response and development of organ failure in ACLF. The roles of lipid mediators in ACLF were assessed by LC-MS in plasma from patients with acute decompensation of cirrhosis with ACLF (n=119) and without ACLF (n=127) and healthy controls (n=25) [18]. Lipid mediators were found to distinguish patients at any stage from healthy controls. For example, leukotriene E4 (LTE4) was found to gradually increase with severity of the disease showing higher levels in ACLF patients with advanced stage disease [49•]. Mucke et al. (2020) conducted targeted LC-MS to investigate sphingosine-1-phosphate (S1P) as biomarker for disease severity in chronic liver disease. S1P levels were found to progressively decrease in patients with acute decompensated cirrhosis to ACLF and were predictive of early mortality [50].
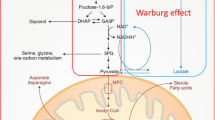
Together, these studies evaluated different potential metabolites for predicting and characterising ACLF development as well as predicting disease severity. In these studies, similar classes of compounds were found to be associated with ACLF. They reported changes in metabolites related to energy metabolism, amino acids, lipids, bile acids, and microbial metabolites (Fig. 1). These metabolites were suggested to be beneficial in predicting ACLF development and associated complications.
Summary of important metabolites and processes which were identified in patients with ACLF. Highlighted in blue metabolites which increased and in red metabolites or process which decreased in blood of ACLF patients. Metabolic alterations were identified in energy-related metabolism, fatty acylcarnitines, lipids, amino acids, and microbial-related metabolites.
HCC: potential biomarkers
Patients with cirrhosis have a high risk for the development of HCC. HCC is the third cause of death related to cancer globally with an approximate 500,000–1,000,000 new cases per year [51]. When HCC is diagnosed early, resection or liver transplantation can be an effective option for treatment [52]. Therefore, there is an increasing effort on finding biomarkers for early diagnosis of HCC given the poor performance of present markers. Metabolites from different compound classes have been defined including amino acids, bile acids, free fatty acids, phospholipids, and LPC (Table 3). Similar classes of compounds were also found to discriminate patients with cirrhosis and ACLF.
In a GC-MS study, analysis was conducted on plasma samples from HCC (n=63) and patient with cirrhosis (n=65). The study demonstrated that combined clinical parameters such as alpha-fetoprotein (AFP) and underlying aetiology and metabolites are effective for early diagnosis of HCC and lead to improved AUC when compared to AFP. The result of their investigation identified amino acids, sugars, fatty acids, and organic acids as important metabolites to distinguish between cirrhosis and HCC. They reported an increase of valine, serine, isoleucine, alpha D-glucosamine 1-phosphate, and linoleic acid [53•]. Plasma phospholipids were shown to have the potential for discriminating between HCC and patients with cirrhosis. Higher phosphatidylcholine (PC), sphingomyelin (SM), and low LPC 20:4 and plasmalogen- phosphatidylethanolamine (pPE) were found in HCC [54]. Lower concentrations of proline and higher level of hydroxyourine were reported in HCC patients compared to patient with cirrhosis [58].
A combination of untargeted GC-MS and targeted LC-MS was used to study serum samples in healthy control (n=50), HCC (n=53), and cirrhosis (n=47) to distinguish between cirrhosis and early HCC. Metabolites related to ammonia recycling, the urea cycle, and amino acid metabolism were found to discriminate between HCC and patient with cirrhosis. A panel of metabolites including methionine, proline, ornithine, pimelylcarnitine, and octanoylcarnitine demonstrated higher AUC (AUC: 0.75) than AFP alone and was validated in another cohort showing reliability of the established model [55]. In an exploratory study, untargeted NMR spectroscopy and targeted LC-MS were conducted to find metabolites for estimating risk of HCC development. Analysis showed that phenylalanine and glutamine were associated with development of HCC in patients with cirrhosis [56].
In a large multicentre study, LC-MS was performed to identify biomarkers for the diagnosis of HCC. A panel of metabolites including phenylalanyl-tryptophan (Phe-Trp) and glycocholate (GCA) was identified and validated in an independent cohort. The identified panel showed a better diagnosis of HCC in high-risk cirrhosis with an AUC of 0.807–0.930. When the panel combined with AFP, the sensitivity was further improved. Furthermore, the specificity of this panel was tested against another two types of cancer with AUC of 0.946 and 0.829 [57•].
Overall, the previous studies focused on finding diagnostic panel of metabolites for early diagnosis of HCC in high-risk patient with cirrhosis (Fig. 2). Although the identified biomarkers showed high specificity and sensitivity and a superior predictive power when compared to AFP in discriminating between HCC and cirrhosis, consistency across multiple studies or appropriate targeted, quantitative assays have not been performed. A large number of multicentre, quantitative measurements, with longitudinal follow-up is required to confirm the clinical utility of the identified biomarkers. At present there are no metabolomic prognostic biomarkers for HCC.
Conclusions and future directions
Metabolomics is a powerful tool for the identification of novel biomarkers for disease diagnosis and progression with particular promise in advanced liver disease. Understanding the significance of altered pathways may aid in the development of better therapeutic approaches. In this review, current research from the past 3 years has been summarised with the aim of identifying metabolites associated with cirrhosis, decompensation/ACLF, and HCC. In cirrhosis, alterations in amino acids, carbohydrates, lipids, bile acids, and microbial metabolites have been identified. Most studies demonstrated a reduction in lipid concentration and increases in bile acids with a shift to increased aromatic amino acid concentrations. Many studies evaluated different potential metabolites for predicting and characterising ACLF development as well as predicting disease severity. In these studies, a similar class of compounds were found to be associated with ACLF with mitochondrial dysfunction associated with systemic inflammation a novel pathway of interest to explore. They reported changes in metabolites related to energy metabolism, amino acids, lipids, bile acids, and microbial metabolites. Patient with cirrhosis have a high risk for developing HCC. Therefore, many studies aimed at finding potential biomarkers to distinguish between the two groups, but these remain elusive. Metabolites including amino acids, bile acids, free fatty acids, phospholipids, and LPC have been identified as important for discrimination between cirrhosis and HCC but agreement between studies is poor.
Although the previous studies have identified many potential metabolites for cirrhosis, ACLF, and HCC, there are some limitations which prevent the generalisation of these results. Most of the performed studies are untargeted analyses and so the analytical parameters employed can vary enormously. Although these approaches have benefits in terms of being unbiased, this can be the reason for the different reported metabolites among the different studies. Different analytical platforms may have been used by these studies which can generate diverse and often incomparable metabolomic data. In some studies, untargeted analysis was performed to find suitable metabolite candidates which were then quantified using targeted analysis with LC-MS. This can be a better approach to identify biological relevant metabolites and then accurately quantify these metabolic alterations. In addition, these studies focused on studying metabolic alterations in certain underlying causes of cirrhosis, such as HCV. This can then be extended to other causes to identify whether the observed alterations are specific to the underlying cause or to the cirrhosis process itself in patients.
In these studies, no specific single metabolite has been identified as an ideal biomarker for the different stages of chronic liver disease, but a panel of metabolites showed that as disease progressed there were similar alterations in core metabolites regardless of the underlying causes. The majority of investigations used serum and plasma to perform their metabolic profiling, as these samples are less invasive than biopsies and they reflect metabolic alteration of endogenous metabolites. The sample size was small in most of these studies which required replication of these studies in larger diverse cohort. Careful selection of the control cohort is significant in biomarker discovery, in order to ensure that any metabolic alterations are specific to the disease under investigation. Most of the studies included healthy controls to serve as reference to the diseased group, whereas more appropriate positive controls such as sepsis (for ACLF) or non-liver cancer (for HCC) would be more informative to determine the specific metabolites of interest rather than discover more generic processes with minimal translational potential. The identified metabolites can form the basis for further validation and studies but currently lack the validation and quantitation needed for application into biochemical laboratories clinical practice.
Nevertheless, these studies have progressed our understanding of metabolic dysfunction in cirrhosis, ACLF, and HCC. However, to confirm the clinical utility of the identified biomarkers, larger multicentre studies with larger cohorts and longer follow-up are needed. Validation in an external cohort is needed to confirm the clinical utility of the identified biomarkers using quantitative assays. A combination of panel of metabolites and clinical parameters is likely to provide a better biomarker than a single metabolite, and processes to account for this require development. Furthermore, the contribution of the identified biomarker to the pathology of cirrhosis and related complication can be investigated further in in vitro and in vivo studies to enhance the understanding of these diseases.
References and Recommended Reading
Sepanlou SG, Safiri S, Bisignano C, Ikuta KS, Merat S, Saberifiroozi M, et al. The global, regional, and national burden of cirrhosis by cause in 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol Hepatol. 2020;5(3):245–66.
Younossi Z, Anstee QM, Marietti M, Hardy T, Henry L, Eslam M, et al. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol. 2018;15(1):11–20.
Axley P, Ahmed Z, Arora S, Haas A, Kuo Y-F, Kamath PS, et al. NASH is the most rapidly growing etiology for acute-on-chronic liver failure-related hospitalization and disease burden in the United States: A Population-Based Study. Liver Transplantation. 2019;25(5):695–705.
Basra S, Anand BS. Definition, epidemiology and magnitude of alcoholic hepatitis. World J Hepatol. 2011;3(5):108–13.
Trebicka J, Fernandez J, Papp M, Caraceni P, Laleman W, Gambino C, et al. The PREDICT study uncovers three clinical courses of acutely decompensated cirrhosis that have distinct pathophysiology. J Hepatol. 2020;73(4):842–54.
Hernaez R, Sola E, Moreau R, Gines P. Acute-on-chronic liver failure: an update. Gut. 2017;66(3):541–53.
Moreau R, Jalan R, Gines P, Pavesi M, Angeli P, Cordoba J, et al. Acute-on-chronic liver failure is a distinct syndrome that develops in patients with acute decompensation of cirrhosis. Gastroenterology. 2013;144(7):1426-37, 37 e1-9.
Arroyo V, Moreau R, Kamath PS, Jalan R, Gines P, Nevens F, et al. Acute-on-chronic liver failure in cirrhosis. Nat Rev Dis Primers. 2016;2:16041.
Tarao K, Nozaki A, Ikeda T, Sato A, Komatsu H, Komatsu T, et al. Real impact of liver cirrhosis on the development of hepatocellular carcinoma in various liver diseases-meta-analytic assessment. Cancer Med. 2019;8(3):1054–65.
Abbiss H, Maker GL, Trengove RD. Metabolomics approaches for the diagnosis and understanding of kidney diseases. Metabolites. 2019;9(2):34.
Rochfort S. Metabolomics reviewed: a new "omics" platform technology for systems biology and implications for natural products research. J Nat Prod. 2005;68(12):1813–20.
Wishart DS. Emerging applications of metabolomics in drug discovery and precision medicine. Nat Rev Drug Discov. 2016;15(7):473–84.
Wishart DS. Metabolomics for Investigating Physiological and Pathophysiological Processes. Physiological Reviews. 2019;99(4):1819–75.
Want EJ, Masson P, Michopoulos F, Wilson ID, Theodoridis G, Plumb RS, et al. Global metabolic profiling of animal and human tissues via UPLC-MS. Nature Protocols. 2013;8(1):17–32.
Mikkonen JJ, Singh SP, Herrala M, Lappalainen R, Myllymaa S, Kullaa AM. Salivary metabolomics in the diagnosis of oral cancer and periodontal diseases. J Periodontal Res. 2016;51(4):431–7.
Pijls KE, Smolinska A, Jonkers DMAE, Dallinga JW, Masclee AAM, Koek GH, et al. A profile of volatile organic compounds in exhaled air as a potential non-invasive biomarker for liver cirrhosis. Scientific Reports. 2016;6(1):19903.
Wishart DS, Lewis MJ, Morrissey JA, Flegel MD, Jeroncic K, Xiong Y, et al. The human cerebrospinal fluid metabolome. Journal of Chromatography B. 2008;871(2):164–73.
Nunes de Paiva MJ, Menezes HC, de Lourdes CZ. Sampling and analysis of metabolomes in biological fluids. Analyst. 2014;139(15):3683–94.
Want EJ. LC-MS untargeted analysis. Methods Mol Biol. 1738;2018:99–116.
Cui L, Lu H, Lee YH. Challenges and emergent solutions for LC-MS/MS based untargeted metabolomics in diseases. Mass Spectrometry Reviews. 2018;37(6):772–92.
Griffiths WJ, Koal T, Wang Y, Kohl M, Enot DP, Deigner H-P. Targeted metabolomics for biomarker discovery. Angewandte Chemie International Edition. 2010;49(32):5426–45.
Emwas AH. The strengths and weaknesses of NMR spectroscopy and mass spectrometry with particular focus on metabolomics research. Methods Mol Biol. 2015;1277:161–93.
Zeki ÖC, Eylem CC, Reçber T, Kır S, Nemutlu E. Integration of GC–MS and LC–MS for untargeted metabolomics profiling. Journal of Pharmaceutical and Biomedical Analysis. 2020;190:113509.
Emwas A-H, Roy R, McKay RT, Tenori L, Saccenti E, Gowda GAN, et al. NMR spectroscopy for metabolomics research. Metabolites. 2019;9(7):123.
Nassar AF, Wu T, Nassar SF, Wisnewski AV. UPLC-MS for metabolomics: a giant step forward in support of pharmaceutical research. Drug Discov Today. 2017;22(2):463–70.
Shackleton C, Pozo OJ, Marcos J. GC/MS in recent years has defined the normal and clinically disordered steroidome: will it soon be surpassed by LC/tandem MS in this role? J Endocr Soc. 2018;2(8):974–96.
Begou O, Gika HG, Wilson ID, Theodoridis G. Hyphenated MS-based targeted approaches in metabolomics. Analyst. 2017;142(17):3079–100.
Gorrochategui E, Jaumot J, Lacorte S, Tauler R. Data analysis strategies for targeted and untargeted LC-MS metabolomic studies: overview and workflow. TrAC Trends in Analytical Chemistry. 2016;82:425–42.
Smolinska A, Blanchet L, Buydens LM, Wijmenga SS. NMR and pattern recognition methods in metabolomics: from data acquisition to biomarker discovery: a review. Anal Chim Acta. 2012;750:82–97.
Xia J, Broadhurst DI, Wilson M, Wishart DS. Translational biomarker discovery in clinical metabolomics: an introductory tutorial. Metabolomics. 2013;9(2):280–99.
Yoo HJ, Jung KJ, Kim M, Kim M, Kang M, Jee SH, et al. Liver cirrhosis patients who had normal liver function before liver cirrhosis development have the altered metabolic profiles before the disease occurrence compared to healthy controls. Front Physiol. 2019;10:1421.
Zheng H, Chen M, Lu S, Zhao L, Ji J, Gao H. Metabolic characterization of hepatitis B virus-related liver cirrhosis using NMR-based serum metabolomics. Metabolomics. 2017;13(10):121.
• Xie G, Wang X, Wei R, Wang J, Zhao A, Chen T, et al. Serum metabolite profiles are associated with the presence of advanced liver fibrosis in Chinese patients with chronic hepatitis B viral infection. BMC Med. 2020;18(1):144 This study used targeted LC-MS to identify alteration in serum metabolites associated with chronic liver disease in HBV-infected patients. The selected predictive metabolites were validated in external cohort.
Salguero S, Rojo D, Berenguer J, Gonzalez-Garcia J, Fernandez-Rodriguez A, Brochado-Kith O, et al. Plasma metabolomic fingerprint of advanced cirrhosis stages among HIV/HCV-coinfected and HCV-monoinfected patients. Liver Int. 2020;40(9):2215–27.
Cano A, Marino Z, Millet O, Martinez-Arranz I, Navasa M, Falcon-Perez JM, et al. A Metabolomics signature linked to liver fibrosis in the serum of transplanted hepatitis C patients. Sci Rep. 2017;7(1):10497.
• Liu Z, Zhang Z, Huang M, Sun X, Liu B, Guo Q, et al. Taurocholic acid is an active promoting factor, not just a biomarker of progression of liver cirrhosis: evidence from a human metabolomic study and in vitro experiments. BMC Gastroenterol. 2018;18(1):112 In this study, targeted LC-MS was conducted to identify the level of bile acid and their contribution to progression of cirrhosis.
Shao L, Ling Z, Chen D, Liu Y, Yang F, Li L. Disorganized gut microbiome contributed to liver cirrhosis progression: a meta-omics-based study. Front Microbiol. 2018;9:3166.
Adams LA, Wang Z, Liddle C, Melton PE, Ariff A, Chandraratna H, et al. Bile acids associate with specific gut microbiota, low-level alcohol consumption and liver fibrosis in patients with non-alcoholic fatty liver disease. Liver Int. 2020;40(6):1356–65.
McPhail MJ, Montagnese S, Villanova M, El Hadi H, Amodio P, Crossey MM, et al. Urinary metabolic profiling by (1)H NMR spectroscopy in patients with cirrhosis may discriminate overt but not covert hepatic encephalopathy. Metab Brain Dis. 2017;32(2):331–41.
Mindikoglu AL, Opekun AR, Putluri N, Devaraj S, Sheikh-Hamad D, Vierling JM, et al. Unique metabolomic signature associated with hepatorenal dysfunction and mortality in cirrhosis. Transl Res. 2018;195:25–47.
Bajaj JS, Fan S, Thacker LR, Fagan A, Gavis E, White MB, et al. Serum and urinary metabolomics and outcomes in cirrhosis. PLoS One. 2019;14(9):e0223061.
Qi SW, Tu ZG, Peng WJ, Wang LX, Ou-Yang X, Cai AJ, et al. (1)H NMR-based serum metabolic profiling in compensated and decompensated cirrhosis. World J Gastroenterol. 2012;18(3):285–90.
Yu M, Zhu Y, Cong Q, Wu C. Metabonomics research progress on liver diseases. Can J Gastroenterol Hepatol. 2017;2017:8467192.
Bajaj JS, Heuman DM, Hylemon PB, Sanyal AJ, White MB, Monteith P, et al. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J Hepatol. 2014;60(5):940–7.
Bajaj JS. Altered microbiota in cirrhosis and its relationship to the development of infection. Clin Liver Dis (Hoboken). 2019;14(3):107–11.
Mahmud N, Kaplan DE, Taddei TH, Goldberg DS. Incidence and mortality of acute-on-chronic liver failure using two definitions in patients with compensated cirrhosis. Hepatology. 2019;69(5):2150–63.
Bajaj JS, Reddy KR, O'Leary JG, Vargas HE, Lai JC, Kamath PS, et al. Serum Levels of metabolites produced by intestinal microbes and lipid moieties independently associated with acute-on-chronic liver failure and death in patients with cirrhosis. Gastroenterology. 2020;159(5):1715–30 e12.
• Moreau R, Claria J, Aguilar F, Fenaille F, Lozano JJ, Junot C, et al. Blood metabolomics uncovers inflammation-associated mitochondrial dysfunction as a potential mechanism underlying ACLF. J Hepatol. 2020;72(4):688–701 This large study from CANONIC demonstrates how mitochondrial dysfunction may underlie the pathophysiology of ACLF.
• Lopez-Vicario C, Checa A, Urdangarin A, Aguilar F, Alcaraz-Quiles J, Caraceni P, et al. Targeted lipidomics reveals extensive changes in circulating lipid mediators in patients with acutely decompensated cirrhosis. J Hepatol. 2020;73(4):817–28 This is a targeted lipidomic study where they associated specific lipid mediators with severity and progression in patients with AD.
Mucke VT, Maria Schwarzkopf K, Thomas D, Mucke MM, Ruschenbaum S, Trebicka J, et al. Serum sphingosine-1-phosphate is decreased in patients with acute-on-chronic liver failure and predicts early mortality. Hepatol Commun. 2020;4(10):1477–86.
Gomaa A-I, Khan S-A, Toledano M-B, Waked I, Taylor-Robinson S-D. Hepatocellular carcinoma: epidemiology, risk factors and pathogenesis. World journal of gastroenterology. 2008;14(27):4300–8.
Kow AWC. Transplantation versus liver resection in patients with hepatocellular carcinoma. Transl Gastroenterol Hepatol. 2019;4:33.
• Di Poto C, Ferrarini A, Zhao Y, Varghese RS, Tu C, Zuo Y, et al. Metabolomic characterization of hepatocellular carcinoma in patients with liver cirrhosis for biomarker discovery. Cancer Epidemiol Biomarkers Prev. 2017;26(5):675–83 In this study, both untargeted and targeted GC-MS analyses were performed to identify metabolites which discriminate between healthy control and HCC. Predicitve models were used to select important metabolites which were evaluated by AUC.
Cotte AK, Cottet V, Aires V, Mouillot T, Rizk M, Vinault S, et al. Phospholipid profiles and hepatocellular carcinoma risk and prognosis in cirrhotic patients. Oncotarget. 2019;10(22):2161–72.
Kim DJ, Cho EJ, Yu K-S, Jang I-J, Yoon J-H, Park T, et al. Comprehensive metabolomic search for biomarkers to differentiate early stage hepatocellular carcinoma from cirrhosis. Cancers. 2019;11(10):1497.
Liang KH, Cheng ML, Lo CJ, Lin YH, Lai MW, Lin WR, et al. Plasma phenylalanine and glutamine concentrations correlate with subsequent hepatocellular carcinoma occurrence in liver cirrhosis patients: an exploratory study. Sci Rep. 2020;10(1):10926.
• Luo P, Yin P, Hua R, Tan Y, Li Z, Qiu G, et al. A large-scale, multicenter serum metabolite biomarker identification study for the early detection of hepatocellular carcinoma. Hepatology. 2018;67(2):662–75 This is a large multicenter study, where they used both untargeted and targeted LC-MS analyses and was validated in external cohort.
Zhou PC, Sun LQ, Shao L, Yi LZ, Li N, Fan XG. Establishment of a pattern recognition metabolomics model for the diagnosis of hepatocellular carcinoma. World J Gastroenterol. 2020;26(31):4607–23.
Availability of data and material
N/A
Code availability
N/A
Author information
Authors and Affiliations
Contributions
Equal
Corresponding author
Ethics declarations
Ethics approval
N/A
Consent to participate
N/A
Consent for publication
N/A
Conflict of interest
Noora Kano declares that he has no conflict of interest. Elizabeth J. Want declares that she has no conflict of interest. Mark JW McPhail declares that he has no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Liver
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kano, N., Want, E.J. & McPhail, M.J.W. Metabolomics in Advanced Liver Disease. Curr Treat Options Gastro 19, 380–397 (2021). https://doi.org/10.1007/s11938-021-00347-w
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11938-021-00347-w