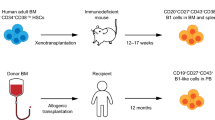
Abstract
In this review we summarize recent insights into the development of human B cells primarily by studying immunodeficiencies. Development and differentiation of B cells can be considered as a paradigm for many other developmental processes in cell biology. However, it differs from the development of many other cell types by phases of extremely rapid cell division and by defined series of somatic recombination and mutation events required to assemble and refine the B cell antigen receptors. Both somatic DNA alteration and proliferation phases take place in defined sites but in different organs. Thus, cell migration and timely arrival at defined sites are additional features of B cell development. By comparing experimental mouse models with insights gained from studying defined genetic defects leading to primary immunodeficiencies and hypogammaglobulinemia, we address important features that are characteristic for human B cells. We also summarize recent advances made by developing improved in vitro and in vivo systems allowing the development of human B cells from hematopoietic stem cells. Combined with genetic and functional studies of immunodeficiencies, these models will contribute not only to a better understanding of disease affecting the B lymphocyte compartment, but also to designing better and safer novel B cell-targeted therapies in autoimmunity and allergy.
Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as : • Of importance •• Of major importance
Ghia P, ten Boekel E, Rolink AG, Melchers F. B-cell development: a comparison between mouse and man. Immunol Today. 1998;19:480–5.
Ferreiros-Vidal I et al. Genome-wide identification of Ikaros targets elucidates its contribution to mouse B-cell lineage specification and pre-B-cell differentiation. Blood. 2013;121:1769–82.
Georgopoulos K. Haematopoietic cell-fate decisions, chromatin regulation and ikaros. Nat Rev Immunol. 2002;2:162–74.
Yoshida T, Ng SY, Zuniga-Pflucker JC, Georgopoulos K. Early hematopoietic lineage restrictions directed by Ikaros. Nat Immunol. 2006;7:382–91.
Thompson EC et al. Ikaros DNA-binding proteins as integral components of B cell developmental-stage-specific regulatory circuits. Immunity. 2007;26:335–44.
Ma S et al. Ikaros and Aiolos inhibit pre-B-cell proliferation by directly suppressing c-Myc expression. Mol Cell Biol. 2010;30:4149–58.
Sun L et al. Expression of dominant-negative and mutant isoforms of the antileukemic transcription factor Ikaros in infant acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 1999;96:680–5.
Holmfeldt L et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet. 2013;45:242–52. This extensive genomic profiling approach to ALL reports chromosomal alterations linked to Ras, Ikaros and TP53.
Molnar A, Georgopoulos K. The Ikaros gene encodes a family of functionally diverse zinc finger DNA-binding proteins. Mol Cell Biol. 1994;14:8292–303.
Papathanasiou P et al. Widespread failure of hematolymphoid differentiation caused by a recessive niche-filling allele of the Ikaros transcription factor. Immunity. 2003;19:131–44.
Schjerven H et al. Selective regulation of lymphopoiesis and leukemogenesis by individual zinc fingers of Ikaros. Nat Immunol. 2013;14:1073–83. The authors demonstrate distinct roles of individual Ikaros Zn-fingers in gene regulation.
Kim J et al. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10:345–55.
Zhang J et al. Harnessing of the nucleosome-remodeling-deacetylase complex controls lymphocyte development and prevents leukemogenesis. Nat Immunol. 2012;13:86–94. The paper describes how Ikaros regulates nucleosome remodeling and histone modification.
Turner Jr CA, Mack DH, Davis MM. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell. 1994;77:297–306.
Ye BH et al. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat Genet. 1997;16:161–70.
Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004;432:635–9.
Kosan C et al. Transcription factor miz-1 is required to regulate interleukin-7 receptor signaling at early commitment stages of B cell differentiation. Immunity. 2010;33:917–28.
Moroy T, Khandanpour C. Growth factor independence 1 (Gfi1) as a regulator of lymphocyte development and activation. Semin Immunol. 2011;23:368–78.
de Almeida Ribeiro C et al. The DNA-binding protein CTCF limits proximal Vkappa recombination and restricts kappa enhancer interactions to the immunoglobulin kappa light chain locus. Immunity. 2011;35:501–13.
Nutt SL, Kee BL. The transcriptional regulation of B cell lineage commitment. Immunity. 2007;26:715–25.
Singh H, Medina KL, Pongubala JM. Contingent gene regulatory networks and B cell fate specification. Proc Natl Acad Sci U S A. 2005;102:4949–53.
Nechanitzky R et al. Transcription factor EBF1 is essential for the maintenance of B cell identity and prevention of alternative fates in committed cells. Nat Immunol. 2013;14:867–75. The paper shows that EBF1 and Pax5 induce lineage commitment by repressing alternative cell fates.
Borghesi L et al. E47 is required for V(D)J recombinase activity in common lymphoid progenitors. J Exp Med. 2005;202:1669–77.
Lin H, Grosschedl R. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature. 1995;376:263–7.
Vallespinos M et al. B Lymphocyte commitment program is driven by the proto-oncogene c-Myc. J Immunol. 2011;186:6726–36.
Seo W, Ikawa T, Kawamoto H, Taniuchi I. Runx1-Cbfbeta facilitates early B lymphocyte development by regulating expression of Ebf1. J Exp Med. 2012;209:1255–62.
Lin YC et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol. 2010;11:635–43.
Zandi S et al. EBF1 is essential for B-lineage priming and establishment of a transcription factor network in common lymphoid progenitors. J Immunol. 2008;181:3364–72.
Welinder E, Ahsberg J, Sigvardsson M. B-lymphocyte commitment: identifying the point of no return. Semin Immunol. 2011;23:335–40.
Dengler HS et al. Distinct functions for the transcription factor Foxo1 at various stages of B cell differentiation. Nat Immunol. 2008;9:1388–98.
Nutt SL, Heavey B, Rolink AG, Busslinger M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401:556–62.
Nutt SL, Eberhard D, Horcher M, Rolink AG, Busslinger M. Pax5 determines the identity of B cells from the beginning to the end of B-lymphopoiesis. Int Rev Immunol. 2001;20:65–82.
Delogu A et al. Gene repression by Pax5 in B cells is essential for blood cell homeostasis and is reversed in plasma cells. Immunity. 2006;24:269–81.
Cobaleda C, Schebesta A, Delogu A, Busslinger M. Pax5: the guardian of B cell identity and function. Nat Immunol. 2007;8:463–70.
Souabni A, Cobaleda C, Schebesta M, Busslinger M. Pax5 promotes B lymphopoiesis and blocks T cell development by repressing Notch1. Immunity. 2002;17:781–93.
Sanz E, Alvarez-Mon M, Martinez AC, de la Hera A. Human cord blood CD34+Pax-5+ B-cell progenitors: single-cell analyses of their gene expression profiles. Blood. 2003;101:3424–30.
Reynaud D, Lefort N, Manie E, Coulombel L, Levy Y. In vitro identification of human pro-B cells that give rise to macrophages, natural killer cells, and T cells. Blood. 2003;101:4313–21.
van Zelm MC et al. Ig gene rearrangement steps are initiated in early human precursor B cell subsets and correlate with specific transcription factor expression. J Immunol. 2005;175:5912–22.
Puck JM et al. The interleukin-2 receptor gamma chain maps to Xq13.1 and is mutated in X-linked severe combined immunodeficiency, SCIDX1. Hum Mol Genet. 1993;2:1099–104.
Prieyl JA, LeBien TW. Interleukin 7 independent development of human B cells. Proc Natl Acad Sci U S A. 1996;93:10348–53.
Puel A, Ziegler SF, Buckley RH, Leonard WJ. Defective IL7R expression in T(-)B(+)NK(+) severe combined immunodeficiency. Nat Genet. 1998;20:394–7.
Campana D, Farrant J, Inamdar N, Webster AD, Janossy G. Phenotypic features and proliferative activity of B cell progenitors in X-linked agammaglobulinemia. J Immunol. 1990;145:1675–80.
Tsukada S et al. Deficient expression of a B cell cytoplasmic tyrosine kinase in human X-linked agammaglobulinemia. Cell. 1993;72:279–90.
Minegishi Y et al. An essential role for BLNK in human B cell development. Science. 1999;286:1954–7.
Mansur A, Therattil J, Young RM, Frieri M. An atypical case of hypogammaglobulinemia. Ann Allergy Asthma Immunol. 2000;84:583–6.
Goldman JP et al. Enhanced human cell engraftment in mice deficient in RAG2 and the common cytokine receptor gamma chain. Br J Haematol. 1998;103:335–42.
van der Loo JC et al. Nonobese diabetic/severe combined immunodeficiency (NOD/SCID) mouse as a model system to study the engraftment and mobilization of human peripheral blood stem cells. Blood. 1998;92:2556–70.
Peled A et al. Dependence of human stem cell engraftment and repopulation of NOD/SCID mice on CXCR4. Science. 1999;283:845–8.
Ueda T et al. Hematopoietic repopulating ability of cord blood CD34(+) cells in NOD/Shi-scid mice. Stem Cells. 2000;18:204–13.
Hogan CJ, Shpall EJ, Keller G. Differential long-term and multilineage engraftment potential from subfractions of human CD34+ cord blood cells transplanted into NOD/SCID mice. Proc Natl Acad Sci U S A. 2002;99:413–8.
Ito M et al. NOD/SCID/gamma(c)(null) mouse: an excellent recipient mouse model for engraftment of human cells. Blood. 2002;100:3175–82.
Traggiai E et al. Development of a human adaptive immune system in cord blood cell-transplanted mice. Science. 2004;304:104–7.
Bente DA, Melkus MW, Garcia JV, Rico-Hesse R. Dengue fever in humanized NOD/SCID mice. J Virol. 2005;79:13797–9.
Gorantla S et al. Human immunodeficiency virus type 1 pathobiology studied in humanized BALB/c-Rag2-/-gammac-/- mice. J Virol. 2007;81:2700–12.
Yu CI et al. Broad influenza-specific CD8+ T-cell responses in humanized mice vaccinated with influenza virus vaccines. Blood. 2008;112:3671–8.
Banerjee P et al. Adult T-cell leukemia/lymphoma development in HTLV-1-infected humanized SCID mice. Blood. 2010;115:2640–8.
Rathinam C et al. Efficient differentiation and function of human macrophages in humanized CSF-1 mice. Blood. 2011;118:3119–28.
Hu Z, Yang YG. Full reconstitution of human platelets in humanized mice after macrophage depletion. Blood. 2012;120:1713–6.
Tanaka S et al. Development of mature and functional human myeloid subsets in hematopoietic stem cell-engrafted NOD/SCID/IL2rgammaKO mice. J Immunol. 2012;188:6145–55.
Kraus H et al. A feeder free differentiation system identifies autonomously proliferating B cell precursors in human bone marrow. J Immunol. 2014;192(3):1044–54. This paper reports a feeder cell-free differentiation system for human B cells and autonomous proliferation of human pro-B and pre-B cells. The paper reports autonomous development and proliferation of human B cell precursors.
Rawlings DJ, Quan SG, Kato RM, Witte ON. Long-term culture system for selective growth of human B-cell progenitors. Proc Natl Acad Sci U S A. 1995;92:1570–4.
Fluckiger AC et al. In vitro reconstitution of human B-cell ontogeny: from CD34(+) multipotent progenitors to Ig-secreting cells. Blood. 1998;92:4509–20.
Kurosaka D, LeBien TW, Pribyl JA. Comparative studies of different stromal cell microenvironments in support of human B-cell development. Exp Hematol. 1999;27:1271–81.
La Motte-Mohs RN, Herer E, Zuniga-Pflucker JC. Induction of T-cell development from human cord blood hematopoietic stem cells by Delta-like 1 in vitro. Blood. 2005;105:1431–9.
Taguchi T et al. Interleukin-7 contributes to human pro-B-cell development in a mouse stromal cell-dependent culture system. Exp Hematol. 2007;35:1398–407.
Ichii M et al. Regulation of human B lymphopoiesis by the transforming growth factor-beta superfamily in a newly established coculture system using human mesenchymal stem cells as a supportive microenvironment. Exp Hematol. 2008;36:587–97.
Awong G et al. Human proT-cells generated in vitro facilitate hematopoietic stem cell-derived T-lymphopoiesis in vivo and restore thymic architecture. Blood. 2013;122:4210–9.
Wardemann H et al. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–7.
Melamed D, Nemazee D. Self-antigen does not accelerate immature B cell apoptosis, but stimulates receptor editing as a consequence of developmental arrest. Proc Natl Acad Sci U S A. 1997;94:9267–72.
Pelanda R et al. Receptor editing in a transgenic mouse model: site, efficiency, and role in B cell tolerance and antibody diversification. Immunity. 1997;7:765–75.
Nemazee DA, Burki K. Clonal deletion of B lymphocytes in a transgenic mouse bearing anti-MHC class I antibody genes. Nature. 1989;337:562–6. A seminal in vivo model to study clonal deletion of autoreactive B cells.
Brombacher F, Kohler G, Eibel H. B cell tolerance in mice transgenic for anti-CD8 immunoglobulin mu chain. J Exp Med. 1991;174:1335–46.
Erikson J et al. Expression of anti-DNA immunoglobulin transgenes in non-autoimmune mice. Nature. 1991;349:331–4.
Hartley SB et al. Elimination from peripheral lymphoid tissues of self-reactive B lymphocytes recognizing membrane-bound antigens. Nature. 1991;353:765–9.
Zikherman J, Parameswaran R, Weiss A. Endogenous antigen tunes the responsiveness of naive B cells but not T cells. Nature. 2012;489:160–4. A very elegant expermental model demonstrating that all peripheral B cells bind autoantigens although with different avidities.
Nemazee D, Hogquist KA. Antigen receptor selection by editing or downregulation of V(D)J recombination. Curr Opin Immunol. 2003;15:182–9.
Hartley SB et al. Elimination of self-reactive B lymphocytes proceeds in two stages: arrested development and cell death. Cell. 1993;72:325–35.
Fields ML, Erikson J. The regulation of lupus-associated autoantibodies: immunoglobulin transgenic models. Curr Opin Immunol. 2003;15:709–17.
Mackay F, Schneider P, Rennert P, Browning J. BAFF AND APRIL: a tutorial on B cell survival. Annu Rev Immunol. 2003;21:231–64.
Frieri M. Mechanisms of disease for the clinician: systemic lupus erythematosus. Ann Allergy Asthma Immunol. 2013;110:228–32.
Claudio E, Brown K, Park S, Wang H, Siebenlist U. BAFF-induced NEMO-independent processing of NF-kappa B2 in maturing B cells. Nat Immunol. 2002;3:958–65.
Zubair A, Frieri M. NF-kappaB and systemic lupus erythematosus: examining the link. J Nephrol. 2013;26:953–9.
Patke A, Mecklenbrauker I, Erdjument-Bromage H, Tempst P, Tarakhovsky A. BAFF controls B cell metabolic fitness through a PKC beta- and Akt-dependent mechanism. J Exp Med. 2006;203:2551–62.
Thompson JS et al. BAFF-R, a newly identified TNF receptor that specifically interacts with BAFF. Science. 2001;293:2108–11.
Sasaki Y, Casola S, Kutok JL, Rajewsky K, Schmidt-Supprian M. TNF family member B cell-activating factor (BAFF) receptor-dependent and -independent roles for BAFF in B cell physiology. J Immunol. 2004;173:2245–52.
Warnatz K et al. B-cell activating factor receptor deficiency is associated with an adult-onset antibody deficiency syndrome in humans. Proc Natl Acad Sci U S A. 2009;106:13945–50.
Pieper K, et al. A common SNP impairs B cell activating factor receptor’s multimerization, contributing to common variable immunodeficiency. J Allergy Clin Immunol. 2014. doi:10.1016/j.jaci.2013.11.021.
Greil J et al. Whole-exome sequencing links caspase recruitment domain 11 (CARD11) inactivation to severe combined immunodeficiency. J Allergy Clin Immunol. 2013;131:1376–83. e1373.
Stepensky P et al. Deficiency of caspase recruitment domain family, member 11 (CARD11), causes profound combined immunodeficiency in human subjects. J Allergy Clin Immunol. 2013;131:477–85. The paper underlines the strict dependece of human B cell development on NF-kappaB signaling.
Schweighoffer E et al. The BAFF Receptor Transduces Survival Signals by Co-opting the B Cell Receptor Signaling Pathway. Immunity. 2013. The authors show that BAFFR-induced signals crossfeed into the BCR pathway and may represent “tonic” BCR signaling.
Yu X et al. Neutralizing antibodies derived from the B cells of 1918 influenza pandemic survivors. Nature. 2008;455:532–6.
Shlomchik MJ, Weisel F. Germinal center selection and the development of memory B and plasma cells. Immunol Rev. 2012;247:52–63.
Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol. 2012;30:429–57.
Lam KP, Kuhn R, Rajewsky K. In vivo ablation of surface immunoglobulin on mature B cells by inducible gene targeting results in rapid cell death. Cell. 1997;90:1073–83.
Liu YJ et al. Mechanism of antigen-driven selection in germinal centres. Nature. 1989;342:929–31.
Liu YJ, Zhang J, Lane PJ, Chan EY, MacLennan IC. Sites of specific B cell activation in primary and secondary responses to T cell-dependent and T cell-independent antigens. Eur J Immunol. 1991;21:2951–62.
Allen CD, Okada T, Tang HL, Cyster JG. Imaging of germinal center selection events during affinity maturation. Science. 2007;315:528–31.
Muramatsu M et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–63.
Revy P et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell. 2000;102:565–75.
Phan TG, Green JA, Gray EE, Xu Y, Cyster JG. Immune complex relay by subcapsular sinus macrophages and noncognate B cells drives antibody affinity maturation. Nat Immunol. 2009;10:786–93.
Okada T et al. Antigen-engaged B cells undergo chemotaxis toward the T zone and form motile conjugates with helper T cells. PLoS Biol. 2005;3:e150.
Pereira JP, Kelly LM, Xu Y, Cyster JG. EBI2 mediates B cell segregation between the outer and centre follicle. Nature. 2009;460:1122–6.
Hannedouche S et al. Oxysterols direct immune cell migration via EBI2. Nature. 2011;475:524–7.
Yi T et al. Oxysterol gradient generation by lymphoid stromal cells guides activated B cell movement during humoral responses. Immunity. 2012;37:535–48.
Kelly LM, Pereira JP, Yi T, Xu Y, Cyster JG. EBI2 guides serial movements of activated B cells and ligand activity is detectable in lymphoid and nonlymphoid tissues. J Immunol. 2011;187:3026–32.
Bannard O et al. Germinal center centroblasts transition to a centrocyte phenotype according to a timed program and depend on the dark zone for effective selection. Immunity. 2013;39:912–24. The paper shows how shuttling between dark and light zone regulates GC B cell proliferation, affinity maturation and memory vs. plasma cell development.
Wang X et al. Follicular dendritic cells help establish follicle identity and promote B cell retention in germinal centers. J Exp Med. 2011;208:2497–510.
Spiegel S, Milstien S. The outs and the ins of sphingosine-1-phosphate in immunity. Nat Rev Immunol. 2011;11:403–15.
Hanel P, Andreani P, Graler MH. Erythrocytes store and release sphingosine 1-phosphate in blood. FASEB J Off Publ Fed Am Soc Exp Biol. 2007;21:1202–9.
Yatomi Y et al. Sphingosine 1-phosphate as a major bioactive lysophospholipid that is released from platelets and interacts with endothelial cells. Blood. 2000;96:3431–8.
Hla T, Venkataraman K, Michaud J. The vascular S1P gradient-cellular sources and biological significance. Biochim Biophys Acta. 2008;1781:477–82.
Venkataraman K et al. Vascular endothelium as a contributor of plasma sphingosine 1-phosphate. Circ Res. 2008;102:669–76.
Pappu R et al. Promotion of lymphocyte egress into blood and lymph by distinct sources of sphingosine-1-phosphate. Science. 2007;316:295–8.
Schwab SR et al. Lymphocyte sequestration through S1P lyase inhibition and disruption of S1P gradients. Science. 2005;309:1735–9.
Matloubian M et al. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature. 2004;427:355–60.
Sic H., et al. S1P-receptors control B cell migration through signaling components associated with primary immunodeficiencies, chronic lymphocytic leukemia and multiple sclerosis. J Allergy Clin Immunol. 2014 in press. The first comprehenisve analysis of S1P-receptor expression and signaling in human B cells.
Green JA, Cyster JG. S1PR2 links germinal center confinement and growth regulation. Immunol Rev. 2012;247:36–51.
MacLennan IC. Germinal centers. Annu Rev Immunol. 1994;12:117–39.
Casamayor-Palleja M, Feuillard J, Ball J, Drew M, MacLennan IC. Centrocytes rapidly adopt a memory B cell phenotype on co-culture with autologous germinal centre T cell-enriched preparations. Int Immunol. 1996;8:737–44.
Batista FD, Iber D, Neuberger MS. B cells acquire antigen from target cells after synapse formation. Nature. 2001;411:489–94.
Schwickert TA et al. A dynamic T cell-limited checkpoint regulates affinity-dependent B cell entry into the germinal center. J Exp Med. 2011;208:1243–52.
Depoil D et al. Immunological synapses are versatile structures enabling selective T cell polarization. Immunity. 2005;22:185–94.
Takahashi Y, Ohta H, Takemori T. Fas is required for clonal selection in germinal centers and the subsequent establishment of the memory B cell repertoire. Immunity. 2001;14:181–92.
Verbeke CS, Wenthe U, Zentgraf H. Fas ligand expression in the germinal centre. J Pathol. 1999;189:155–60.
Phan TG et al. High affinity germinal center B cells are actively selected into the plasma cell compartment. J Exp Med. 2006;203:2419–24.
Thiel J et al. Genetic CD21 deficiency is associated with hypogammaglobulinemia. J Allergy Clin Immunol. 2012;129:801–10. e806.
van Zelm MC et al. An antibody-deficiency syndrome due to mutations in the CD19 gene. N Engl J Med. 2006;354:1901–12.
van Zelm MC et al. CD81 gene defect in humans disrupts CD19 complex formation and leads to antibody deficiency. J Clin Invest. 2010;120:1265–74.
Tangye SG, Avery DT, Deenick EK, Hodgkin PD. Intrinsic differences in the proliferation of naive and memory human B cells as a mechanism for enhanced secondary immune responses. J Immunol. 2003;170:686–94.
Wang LC et al. The TEL/ETV6 gene is required specifically for hematopoiesis in the bone marrow. Genes Dev. 1998;12:2392–402.
Hymowitz SG et al. Structures of APRIL-receptor complexes: like BCMA, TACI employs only a single cysteine-rich domain for high affinity ligand binding. J Biol Chem. 2005;280:7218–27.
Yuan ZL, Guan YJ, Chatterjee D, Chin YE. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307:269–73.
Kienzler AK, Rizzi M, Reith M, Nutt SL, Eibel H. Inhibition of human B-cell development into plasmablasts by histone deacetylase inhibitor valproic acid. J Allergy Clin Immunol. 2013;131:1695–9.
Yamaguchi T et al. Histone deacetylases 1 and 2 act in concert to promote the G1-to-S progression. Genes Dev. 2010;24:455–69.
Matthias P, Rolink AG. Transcriptional networks in developing and mature B cells. Nat Rev Immunol. 2005;5:497–508.
O’Riordan M, Grosschedl R. Coordinate regulation of B cell differentiation by the transcription factors EBF and E2A. Immunity. 1999;11:21–31.
Horcher M, Souabni A, Busslinger M. Pax5/BSAP maintains the identity of B cells in late B lymphopoiesis. Immunity. 2001;14:779–90.
Lin Y, Wong K, Calame K. Repression of c-myc transcription by Blimp-1, an inducer of terminal B cell differentiation. Science. 1997;276:596–9.
Angelin-Duclos C, Cattoretti G, Lin KI, Calame K. Commitment of B lymphocytes to a plasma cell fate is associated with Blimp-1 expression in vivo. J Immunol. 2000;165:5462–71.
Lin KI, Angelin-Duclos C, Kuo TC, Calame K. Blimp-1-dependent repression of Pax-5 is required for differentiation of B cells to immunoglobulin M-secreting plasma cells. Mol Cell Biol. 2002;22:4771–80.
Shaffer AL et al. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17:51–62.
Shapiro-Shelef M et al. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 2003;19:607–20.
Kallies A et al. Plasma cell ontogeny defined by quantitative changes in blimp-1 expression. J Exp Med. 2004;200:967–77.
Shaffer AL et al. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21:81–93.
Klein U et al. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol. 2006;7:773–82.
Kallies A et al. Initiation of plasma-cell differentiation is independent of the transcription factor Blimp-1. Immunity. 2007;26:555–66.
Cattoretti G et al. BCL-6 protein is expressed in germinal-center B cells. Blood. 1995;86:45–53.
Allman D et al. BCL-6 expression during B-cell activation. Blood. 1996;87:5257–68.
Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276:589–92.
Basso K et al. Integrated biochemical and computational approach identifies BCL6 direct target genes controlling multiple pathways in normal germinal center B cells. Blood. 2010;115:975–84.
Tunyaplin C et al. Direct repression of prdm1 by Bcl-6 inhibits plasmacytic differentiation. J Immunol. 2004;173:1158–65.
Shaffer AL et al. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity. 2000;13:199–212.
Abecasis GR et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. The report of the 1,000 genome project.
Saito M et al. BCL6 suppression of BCL2 via Miz1 and its disruption in diffuse large B cell lymphoma. Proc Natl Acad Sci U S A. 2009;106:11294–9.
Shapiro-Shelef M, Lin KI, Savitsky D, Liao J, Calame K. Blimp-1 is required for maintenance of long-lived plasma cells in the bone marrow. J Exp Med. 2005;202:1471–6.
Reimold AM et al. Transcription factor B cell lineage-specific activator protein regulates the gene for human X-box binding protein 1. J Exp Med. 1996;183:393–401.
Iwakoshi NN et al. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat Immunol. 2003;4:321–9.
Todd DJ et al. XBP1 governs late events in plasma cell differentiation and is not required for antigen-specific memory B cell development. J Exp Med. 2009;206:2151–9.
Mittrucker HW et al. Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science. 1997;275:540–3.
Sciammas R et al. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity. 2006;25:225–36.
Berberich I, Shu GL, Clark EA. Cross-linking CD40 on B cells rapidly activates nuclear factor-kappa B. J Immunol. 1994;153:4357–66.
Han S et al. Cellular interaction in germinal centers. Roles of CD40 ligand and B7-2 in established germinal centers. J Immunol. 1995;155:556–67.
Basso K et al. Tracking CD40 signaling during germinal center development. Blood. 2004;104:4088–96.
Saito M et al. A signaling pathway mediating downregulation of BCL6 in germinal center B cells is blocked by BCL6 gene alterations in B cell lymphoma. Cancer cell. 2007;12:280–92.
Ferrari S et al. Mutations of CD40 gene cause an autosomal recessive form of immunodeficiency with hyper IgM. Proc Natl Acad Sci U S A. 2001;98:12614–9.
Jain A et al. Specific missense mutations in NEMO result in hyper-IgM syndrome with hypohydrotic ectodermal dysplasia. Nat Immunol. 2001;2:223–8.
Good KL, Bryant VL, Tangye SG. Kinetics of human B cell behavior and amplification of proliferative responses following stimulation with IL-21. J Immunol. 2006;177:5236–47.
Ding BB, Bi E, Chen H, Yu JJ, Ye BH. IL-21 and CD40L synergistically promote plasma cell differentiation through upregulation of Blimp-1 in human B cells. J Immunol. 2013;190:1827–36.
Ozaki K et al. Regulation of B cell differentiation and plasma cell generation by IL-21, a novel inducer of Blimp-1 and Bcl-6. J Immunol. 2004;173:5361–71.
Pene J et al. Cutting edge: IL-21 is a switch factor for the production of IgG1 and IgG3 by human B cells. J Immunol. 2004;172:5154–7.
Kotlarz D et al. Loss-of-function mutations in the IL-21 receptor gene cause a primary immunodeficiency syndrome. J Exp Med. 2013;210:433–43. The paper shows the role of IL-21R signaling in immune responses of B, T and NK cells.
Scheeren FA et al. STAT5 regulates the self-renewal capacity and differentiation of human memory B cells and controls Bcl-6 expression. Nat Immunol. 2005;6:303–13.
Diehl SA et al. STAT3-mediated up-regulation of BLIMP1 Is coordinated with BCL6 down-regulation to control human plasma cell differentiation. J Immunol. 2008;180:4805–15.
Avery DT et al. B cell-intrinsic signaling through IL-21 receptor and STAT3 is required for establishing long-lived antibody responses in humans. J Exp Med. 2010;207:155–71.
Holland SM et al. STAT3 mutations in the hyper-IgE syndrome. N Engl J Med. 2007;357:1608–19.
Minegishi Y et al. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature. 2007;448:1058–62.
Hsu AP et al. Intermediate phenotypes in patients with autosomal dominant hyper-IgE syndrome caused by somatic mosaicism. J Allergy Clin Immunol. 2013;131:1586–93.
Compliance with Ethics Guidelines
Conflict of Interest
This work was supported by the German Federal Ministry of Education and Research (BMBF 01 EO 0803) to Marta Rizzi, the German Cancer research fund through grant 109138 and the DFG through the TRR130 to Hermann Eibel and Helene Kraus; Anne-Kathrin Kienzler was supported by the NIHR Oxford Biomedical Research Centre.
Heiko Sic declares that he has no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with animal subjects performed by any of the authors. With regard to the authors’ research cited in this paper, all procedures were followed in accordance with the ethical standards of the responsible committee on human experimentation and with the Helsinki Declaration of 1975, as revised in 2000 and 2008.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is part of the Topical Collection on Basic and Applied Science
Rights and permissions
About this article
Cite this article
Eibel, H., Kraus, H., Sic, H. et al. B cell Biology: An Overview. Curr Allergy Asthma Rep 14, 434 (2014). https://doi.org/10.1007/s11882-014-0434-8
Published:
DOI: https://doi.org/10.1007/s11882-014-0434-8