Abstract
Background
Primary open-angle glaucoma (POAG) is affected by both genetics and environmental factors. CDKN2B-AS1 polymorphisms have been reported to be involved in the pathogenesis of POAG. However, the results of the genetic associations between the CDKN2B-AS1 polymorphisms and POAG risk were inconclusive.
Aims
This study aimed to evaluate the correlation of CDKN2B-AS1 polymorphisms and POAG susceptibility using a meta-analysis.
Methods
Meta-analysis was performed by searching PubMed, Web of science, the Cochrane database of system reviews, CNKI, and Embase databases. The relationship of CDKN2B-AS1 rs4977756, rs10120688, rs2157719, and rs7049105 polymorphisms and POAG risk was evaluated by the odds ratios (ORs) and 95% confidence intervals (CIs).
Results
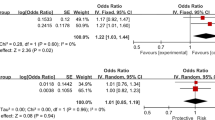
Eleven studies with 8290 cases and 13,485 controls were included in the present meta-analysis. The alleles of rs4977756 and rs10120688 significantly increased the risk of POAG (rs4977756: OR = 1.20, 95%CI = 1.03–1.39, p = 0.02; rs10120688: OR = 1.36, 95%CI = 1.29–1.44, p < 0.00001). As for ethnicity, rs4977756 polymorphism significantly increased POAG risk in Caucasians (OR = 1.33, 95%CI = 1.12–1.57, p = 0.0009), but not in Asians. In addition, the rs2157719 allele was significantly associated with POAG risk in Asians (OR = 0.66, 95%CI = 0.55–0.80, p < 0.0001), but not in Caucasians (p > 0.05).
Conclusions
The CDKN2B-AS1 rs4977756 might increase the POAG risk in Caucasian population, and rs2157719 might decrease the POAG risk in Asian population, while rs10120688 might increase the risk of POAG.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glaucoma is one of the leading causes of blindness worldwide, especially in South Africa characterized by progressive damage of retinal ganglion cells, optic nerve head excavation, and visual field loss [1,2,3,4,5]. Primary open-angle glaucoma (POAG) is one of the most common type of glaucoma with a 4- to fivefold higher risk in South African than that in Caucasian populations [6, 7]. Although the POAG represents the most prevalent form of glaucoma, the pathogenesis and factors determining the disease progression are poorly understood. Risk factors including increased age, elevated intraocular pressure (IOP), African ancestry, and family history were identified in previous studies [8,9,10]. However, the underlying causes of these risk factors remain obscure. Additionally, the genetic factors have illustrated to play an important role in the development of POAG. Numbers of genome-wide association studies (GWAS) on the genetic association of POAG have identified multiple genomic loci in 7q31.1 (caveolin 1 (CAV1)/caveolin 2 (CAV2)) [11, 12], 1q24.1 (transmembrane and coiled-coil domains 1, TMCO1) [13], 14q23 (sin oculis homeobox 1/sin oculis homeobox 6, SIX1/SIX6) [14], and 9p21.3 (cyclin-dependent kinase inhibitor 2B antisense noncoding RNA, CDKN2B-AS1) [15] and genes including cytochrome P450 family 1 subfamily B polypeptide 1 (CYP1B1) [16], WD repeat domain 36 (WDR36) [17], TANK-binding kinase 1 (TBK1) [18], and galactosylceramidase (GALC) [19] in African, Caucasian, and Asian populations.
CDKN2B-AS1 is an antisense RNA that may influence the nearby CDKN2A and CDKN2B genes via regulatory mechanisms [20] and was determinate to be a genetic susceptibility locus for several age-related complex diseases including POAG [21, 22]. Recently, several GWA studies have identified a number of POAG-associated single-nucleotide polymorphisms (SNPs) such as rs4977756, rs10120688, rs2157719, and rs7049105 [23, 24]. Burdon et al. revealed CDKN2B-AS1 rs4977756 can significantly increase the risk of POAG in a cohort of 590 cases and 3956 controls in Australia [23]. Ng et al. also illustrated the genetic association between the CDKN2B-AS1 rs4977756 and POAG in Australians [25]. It is suggested that the stronger genetic signals at the 9p21 locus among females may contribute to the observed sex bias for POAG. However, several studies on the association between CDKN2B-AS1 rs4977756 and POAG in Chinese [26], Japanese [27], Pakistan [28], and American Caucasian [29] populations have shown inconsistent results. For limited studies and relatively small sample size in the correlation of CDKN2B-AS1 rs10120688, rs2157719, and rs7049105 and POAG, inconclusive results were observed.
Meta-analysis is powerful to obtain a more precise conclusion that was inconclusive in previous individual study. Considering the inconclusive results and limited sample size in previous studies, we performed a meta-analysis to further evaluate the genetic associations between CDKN2B-AS1 rs4977756, rs10120688, rs2157719, and rs7049105 polymorphisms and the susceptibility of POAG.
Materials and methods
Search strategy
The relevant literatures published as of April 1, 2021 were searched in PubMed, Chinese National Knowledge Infrastructure (CNKI), CQVIP, Chinese Biomedical Literature Database (CBM), Web of science, Embase, and Wanfang database. “cyclin-dependent kinase inhibitor 2B antisense RNA 1” or “CDKN2B-AS1” and “polymorphism” or “single nucleotide polymorphisms” or “SNPs” and “Primary Open-Angle Glaucoma” or “Open-Angle Glaucoma” or “POAG” or “OAG” were used as search terms. At the same time, the references included and related reviews were reviewed. Language is unlimited.
Inclusion and exclusion criteria
The following criteria were used for the literature inclusion: (1) papers should concern CDKN2BAS polymorphism and glaucoma risk, (2) case–control and/or cohort designed studies, (3) contained SNP genotype data both in case and control groups, (4) adequate data for the calculation of odds ratios (ORs) and 95% confidence intervals (CIs), (5) the genotype distribution in control groups was in Hardy–Weinberg equilibrium (HWE). In addition, studies were excluded when they were (1) studies that contained overlapping data with other literatures; (2) data came from case-reports, reviews, or abstracts; (3) not case–control and/or cohort designed studies; (4) genotype frequencies were unavailable; and (5) the control group did not confirm to HWE.
Data extraction
Liu SS and Chen SW independently extracted all data from each eligible study. Disputes was resolved by discussion. The following information from each study were extracted: the name of first author, publication, ethnicity, mean age, gender, type of glaucoma, mean intraocular pressure (IOP) (mm Hg), mean vertical cup-to-disc ratio (VCDR), and the number of cases and controls.
Statistical analysis
Data was processed with RevMan 5 (Oxford, UK) and STATA12.0. The association between CDKN2B-AS1 polymorphisms and POAG risk was evaluated by pooled OR) and 95%CI. The significance of the pooled OR was assessed by the Z test. I2 was used to evaluate the heterogeneity between studies. If I2 was less than 50% (p > 0.05), the combined effect value OR and its 95%CI were calculated using the fixed-effect model; otherwise, the random effects model was used. Subgroup analyses were conducted based on ethnicity. Egger’s test and Begg’s test were used to evaluate and analyze the publication bias of each study. Meta-regression analysis was used to analyze the potential sources of inter-study heterogeneity. p value < 0.05 was considered to be significant difference.
Results
The characters of eligible studies
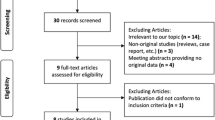
After an exhaustive search, a total of 2154 articles were retrieved from electric databases. According to the inclusion and exclusion criteria, we excluded 507 duplicated studies, 382 not original studies, and 1244 not case–control designed studies (Fig. 1). Finally, 11 studies with 8290 cases and 13,485 controls were enrolled in the present meta-analysis [15, 20, 23, 25,26,27,28,29, 39,40,41]. Among these articles, 7 studies refer to rs4977756, 3 studies refer to rs10120688 and rs2157719 respectively, and 2 studies refer to rs7049105. Furthermore, 4 studies were conducted on Asian populations, and 7 were on Caucasian populations. The basic information and genotype distribution of the included literatures are shown in Table 1.
Results of meta-analysis
The results of the meta-analysis preformed in the current study are presented in Table 2. The allelic models of rs4977756 and rs10120688 significantly increased the risk of POAG (rs4977756: OR = 1.20, 95%CI = 1.03–1.39, p = 0.02; rs10120688: OR = 1.36, 95%CI = 1.29–1.44, p < 0.00001). Subgroup analysis based on ethnicity revealed that the allelic model of rs4977756 significantly increased the POAG risk in Caucasians (OR = 1.33, 95%CI = 1.12–1.57, p = 0.0009), but not in Asians (OR = 1.06, 95%CI = 0.94–1.19, p = 0.34). In addition, the allelic model of rs2157719 was associated with decreased risk of POAG in Asian population (OR = 0.66, 95%CI = 0.55–0.80, p < 0.0001), but not in Caucasian population (OR = 0.97, 95%CI = 0.51–1.84, p = 0.93). Subgroup analysis based on ethnicity was canceled for lack of sufficient data in rs10120688 and rs7049105 (Fig. 2). Moreover, the relationship of the dominant and recessive models of the CDKN2B-AS1 rs4977756, rs10120688, rs2157719, and rs7049105 polymorphisms and POAG susceptibility was canceled for lack of data.
Heterogeneity
Heterogeneity was found for the rs4977756 (I2% = 82, p < 0.0001), rs2157719 (I2% = 93, p < 0.00001), and rs7049105 (I2% = 95, p < 0.00001) in overall analysis. Significant heterogeneity was detected in subgroup analysis based on ethnicity (Caucasians: rs4977756: I2% = 77, p = 0.005; rs2157719: I2% = 93, p = 0.002). The significant heterogeneity in rs4977756 was primarily presented by Burdon et al. and Ng et al. No significant heterogeneity was detected after removal these studies (I2 = 37%, p > 0.05). Moreover, the significant heterogeneity in rs2157719 was primarily presented by Zanon-Moreno et al. And no significant heterogeneity was detected after removal these studies (I2 = 0%, p > 0.05) (Table 2).
Sensitive analysis and publish bias
There was no deviation from Hardy–Weinberg equilibrium or OR 3.0 in the 11 studies on CDKN2B-AS1 rs4977756, rs10120688, rs2157719, and rs7049105 polymorphisms in the present meta-analysis. Thus, the association between CDKN2B-AS1 rs4977756, rs10120688, rs2157719, and rs7049105 and POAG was stable and reliable. As shown in Table 3 and Figs. 3 and 4, no evidence of publication bias was found by using Begg’ s test and Egger’ s test.
Discussion
In the present study, we have assessed the genetic association of CDKN2B-AS1 rs4977756, rs10120688, rs2157719, and rs7049105 and the risk of POAG and found that the CDKN2B-AS1 rs4977756 and rs10120688 can significantly increase the risk of POAG. However, the CDKN2B-AS1 rs4977756 was associated with POAG risk only in Caucasian, but not in Asian. In addition, the rs2157719 decreases the POAG risk only in Asian. No association between rs7049105 and the risk of POAG was detected.
GWAS data have identified significant associations between several genes in chromosome 9p21 and multiple common diseases [30,31,32]. CDKN2B-AS1 is a non-coding gene (also known as ANRIL) with an unknown function, located on chromosome 9p21.3 [33]. Multiple tissue-specific splice variants of this gene have been reported. This gene is likely to play a role in regulating expression of genes through epigenetic mechanisms. CDKN2B-AS1 polymorphisms were illustrated to be associated with coronary artery disease (CAD) [34] and type 2 diabetes mellitus risk initially [35]. Subsequently, the polymorphisms in CDKN2B-AS1 were identified to be significantly associated with the development of POAG [29]. However, little is known about the biological meanings underlying this locus. Song et al. have shown that 33 enhancers were identified in 9p21, some of which being within CDKN2B-AS1 [36]. Thus, it is suggested that the variants in this gene would probably affect the expression level of the downstream genes CDKN2A and CDKN2B, which is response to the elevated IOP in glaucoma. Altered gene expression may also affect cell cycle regulation and lead to a tendency toward apoptosis of retinal ganglion cells. The cells of POAG patients carrying the risk alleles of CDKN2B-AS1 gene may be more sensitive to IOP. Moreover, the variants in this gene might also affect the distant genes that correlated with the complex pathogenesis of glaucoma.
The rs4977756 is mapped 59 kb telomeric to CDKN2B within a 122-kb region of LD at 9p21.3, which encompasses the CDKN2A-CDKN2B tumor suppressor genes and was shown to be associated with the risk of glioma [37, 38]. In this meta-analysis, a total of 5144 cases and 11,285 controls from 7 publications were included. And the CDKN2B-AS1 rs4977756 polymorphism was significantly associated with increased POAG risk. To our knowledge, this is the first time a significant genetic association between CDKN2B-AS1 rs4977756 polymorphism and the susceptibility of POAG was detected using a meta-analysis. As for ethnicity, rs4977756 polymorphism was associated with increased risk of POAG in Caucasians, but not in Asians. Interestingly, GWAS data has demonstrated that the rs4977756 polymorphism was a risk factor for POAG in Japanese, which may suggest that the CDKN2B-AS1 locus in previous study using a Japanese population seemed to be shared with the Caucasian subjects, but not with the Chinese and other Asian populations. Thus, the CDKN2B-AS1 rs4977756 for POAG is still ethnicity related.
The current meta-analysis had various limitations. Firstly, the ethnicity included in the present meta-analysis was relatively limited. Subgroup analysis was only divided into Caucasian and Asian. Considering the significant influence of African ancestry in the pathogenesis of POAG, studies on the association between CDKN2B-AS1 polymorphisms and POAG risk in other ethnicities such as African and Latin American are necessary in the future. Secondly, age and gender were shown to be involved in the pathogenesis of POAG. However, we could not perform further stratification analyses based on age and gender due to limited data. Thirdly, we failed to assess the potential influence of genetic and environmental factors, as well as the gene–gene and gene-environment interactions on POAG for lack of relevant data. Fourthly, CDKN2B-AS1 rs4977756, rs10120688, rs2157719, and rs7049105 polymorphisms would not be enough to explain the associations between CDKN2B-AS1 gene and POAG risk. Fifthly, the dominant and recessive models of these polymorphisms were not available for lack of relevant data.
Conclusions
The CDKN2B-AS1 rs4977756 might increase the POAG risk in Caucasian population. rs10120688 might be associated with an increased risk of POAG. rs2157719 might decrease the risk of POAG in Asian population. To confirm these results, a larger number of subjects with different ethnicity are necessary to explore the role of CDKN2B-AS1 rs4977756 and rs10120688 in POAG.
Availability of data and material
The data used to support the founding of this study are included within the article.
Code availability
Not applicable.
References
Evangelho K, Mogilevskaya M, Losada-Barragan M et al (2017) Pathophysiology of primary open-angle glaucoma from a neuroinflammatory and neurotoxicity perspective: a review of the literature. Int Ophthalmol 10:1–13. https://doi.org/10.1007/s10792-017-0795-9
Wójcik-Gryciuk A, Skup M, Waleszczyk WJ (2015) Glaucoma–state of the art and perspectives on treatment. Restor Neurol Neurosci 34(1):107–123. https://doi.org/10.3233/RNN-150599
Gassel CJ, Reinehr S, Gomes SC et al (2020) Preservation of optic nerve structure by complement inhibition in experimental glaucoma. Cell Tissue Res 382:2835–2848. https://doi.org/10.1007/s00441-020-03240-7
Zhi WL, Chee ML, Thakur S et al (2020) Albuminuria and primary open-angle glaucoma: the Singapore Chinese Eye Study (SCES). Br J Ophthalmol 2020:315920. https://doi.org/10.1136/bjophthalmol-2020-315920
Singh MK (2020) Challenges in diagnosis and management of glaucoma: Indian scenario. Indian Journal of Clinical and Experimental Ophthalmology 6(1):3–4. https://doi.org/10.18231/j.ijceo.2020.002
Pwm B, Cook C, Nag A et al (2017) Genetic African ancestry is associated with central corneal thickness and intraocular pressure in primary open-angle glaucoma. Invest Ophthalmol Vis Sci 58(7):3172–3180. https://doi.org/10.1167/iovs.17-21716
Bs A, Ahm FA, Acvv A et al (2021) Ocular blood flow as it relates to race and disease on glaucoma. Advances in Ophthalmology and Optometry 6:245–262. https://doi.org/10.1016/j.yaoo.2021.04.016
Antar H, Tsikata E, Ratanawongphaibul K et al (2019) Analysis of neuroretinal rim by age, race, and sex using high-density 3-dimensional spectral-domain optical coherence tomography. J Glaucoma 28(11):979–988. https://doi.org/10.1097/IJG.0000000000001381
Marjanović I, Martinez A, Marjanović M et al (2014) Changes in the retrobulbar hemodynamic parameters after decreasing the elevated intraocular pressure in primary open-angle glaucoma patients. Srp Arh Celok Lek 142(5–6):286–290. https://doi.org/10.2298/sarh1406286m
Yutao L, Hauser MA, Akafo SK et al (2013) Investigation of known genetic risk factors for primary open angle glaucoma in two populations of African ancestry. Invest Ophthalmol Vis Sci 54(9):6248–6254. https://doi.org/10.1167/iovs.13-12779
Wiggs JL, Kang JH, Yaspan BL et al (2011) Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma in Caucasians from the USA. Hum Mol Genet 20(23):4707–4713. https://doi.org/10.1093/hmg/ddr382
Huang WB, Wei WM, Zhou MW et al (2013) Association of SNP rs4236601 near CAV1 and CAV2 with primary open-angle glaucoma: a meta-analysis. Clin Exp Ophthalmol 42(6):515–521. https://doi.org/10.1111/ceo.12201
Kondkar AA, Mousa A, Azad TA et al (2016) Polymorphism rs7555523 intransmembrane and coiled-coil domain 1(TMCO1) is not a risk factor for primary open angle glaucoma in a Saudi cohort. J Negat Results Biomed 15(1):17. https://doi.org/10.1186/s12952-016-0060-1
Kondkar AA, Azad TA, Almobarak FA et al (2017) Polymorphism rs10483727 in the SIX1/SIX6 gene locus is a risk factor for primary open angle glaucoma in a Saudi cohort. Genet Test Mol Biomarkers 22(1):74–78. https://doi.org/10.1186/s12952-016-0060-1
Burdon KP, Macgregor S, Hewitt AW et al (2011) Genome-wide association study identifies susceptibility loci for open angle glaucoma at TMCO1 and CDKN2B-AS1. Nat Genet 43(6):574–578. https://doi.org/10.1038/ng.824
Emamalizadeh B, Daneshmandpour Y, Kazeminasb S et al (2021) Mutational analysis of CYP1B1 gene in Iranian pedigrees with glaucoma reveals known and novel mutations. Int Ophthalmol 543–554:1–8. https://doi.org/10.1007/s10792-021-01888-w
Fanny M, Yannick B, Paul CJ et al (2020) Variants of WDR36 in Cameroonian glaucoma patients. Biomed Sci Eng 6(1):043–047. https://doi.org/10.17352/abse.000021
Awadalla MS, Fingert JH, Roos BE et al (2015) Copy number variations of TBK1 in Australian patients with primary open-angle glaucoma. Am J Ophthalmol 159(1):124-130.e1. https://doi.org/10.1016/j.ajo.2014.09.044
Liu Y, Garrett ME, Yaspan BL et al (2014) DNA copy number variants of known glaucoma genes in relation to primary open-angle glaucoma. Invest Ophthalmol Vis 55(12):8251–8258. https://doi.org/10.1167/iovs.14-15712
Burdon KP, Crawford A, Casson RJ et al (2012) Glaucoma risk alleles at CDKN2B-AS1 are associated with lower intraocular pressure, normal-tension glaucoma, and advanced glaucoma. Ophthalmology 119(8):1539–1545. https://doi.org/10.1016/j.ophtha.2012.02.004
Pasquale LR, Loomis SJ, Kang JH et al (2013) CDKN2B-AS1 genotype-glaucoma feature correlations in primary open-angle glaucoma patients from the United States. Am J Ophthalmol 155(2):342–353. https://doi.org/10.1016/j.ajo.2012.07.023
Sayed AA, Al-Nafie AN, Abdullah AS et al (2016) Intronic polymorphisms in theCDKN2B-AS1 gene are strongly associated with the risk of myocardial infarction and coronary artery disease in the Saudi population. Int J Mol Sci 17(3):395. https://doi.org/10.3390/ijms17030395
Burdon KP, Mitchell P, Lee A et al (2015) Association of open-angle glaucoma loci with incident glaucoma in the Blue Mountains Eye study. Am J Ophthalmol 159(1):31–36. https://doi.org/10.1016/j.ajo.2014.09.020
Nakano M, Ikeda Y, Tokuda Y et al (2012) Common variants in CDKN2B-AS1 associated with optic-nerve vulnerability of glaucoma identified by genome-wide association studies in Japanese. PLoS One 7(3):e33389. https://doi.org/10.1371/journal.pone.0033389
Ng SK, Burdon KP, Fitzgerald JT et al (2016) Genetic association at the 9p21 glaucoma locus contributes to sex bias in normal-tension glaucoma. Invest Ophthalmol Vis Sci 57(7):3416–3621. https://doi.org/10.1167/iovs.16-19401
Chen Y, Hughes G, Chen X et al (2015) Genetic variants associated with different risks for high tension glaucoma and normal tension glaucoma in a Chinese population. Invest Ophthalmol Vis Sci 56(4):2595–2600. https://doi.org/10.1167/iovs.14-16269
Kimura Y, Akagi T, Miyake M et al (2015) Association between the CDKN2B-AS1 gene and primary open angle glaucoma with high myopia in Japanese patients. Ophthal Genet 37(2):1–3. https://doi.org/10.3109/13816810.2015.1020559
Micheal S, Ayub H, Khan MI et al (2014) Association of known common genetic variants with primary open angle, primary angle closure, and pseudoexfoliation glaucoma in Pakistani cohorts. Mol Vis 20(20):1471–1479
Cao D, Jiao XD, Liu X et al (2012) CDKN2B Polymorphism is associated with primary open-angle glaucoma (POAG) in the Afro-Caribbean population of Barbados. West Indies PLoS One 7(6):e39278. https://doi.org/10.1371/journal.pone.0039278
Cakmak HA, Bayoğlu B, Durmaz E et al (2015) Evaluation of association between common genetic variants on chromosome 9p21 and coronary artery disease in Turkish population. Anatol J Cardiol 15(3):196–203. https://doi.org/10.5152/akd.2014.5285
Alsaad R, Elmansoury J, Alhazzaa S et al (2020) Chromosome 1q terminal deletion and congenital glaucoma: a case report. American Journal of Case Reports 21:e918128. https://doi.org/10.12659/AJCR.918128
Ng SK, Casson RJ, Burdon KP et al (2014) Chromosome 9p21 primary open-angle glaucoma susceptibility locus: a review. Clin Exp Ophthalmol 42(1):25–32. https://doi.org/10.1111/ceo.12234
Aarabi G, Zeller T, Heydecke G et al (2018) Roles of the Chr.9p21.3ANRILLocus in regulating inflammation and implications for anti-inflammatory drug target identification. Front Cardiovasc Med 5: 47. https://doi.org/10.3389/fcvm.2018.00047
Nawaz SK, Noreen A, Rani A et al (2015) Association of the rs10757274 SNP with coronary artery disease in a small group of a Pakistani population. Anatol J Cardiol 15(9):709–715. https://doi.org/10.5152/akd.2014.5470
Liu ZK, Jian-Ping LI, Zhang Y (2016) Pleiotropic effects of genotypes at rs10757274 locus of coronary heart disease susceptibility gene CDKN2B-AS1 on type 2 diabetes mellitus and hyperlipemia. Chin J Preven Control Chronic Dis 2:158–160
Song X, Rahim NG, Tanasa B et al (2011) 9p21 DNA variants associated with coronary artery disease impair interferon-γ signalling response. Nature 470(7333):264–268. https://doi.org/10.1038/nature09753
Wrensch M, Jenkins RB, Chang JS et al (2009) Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat Genet 41(8):905–908. https://doi.org/10.1038/ng.408
Lu H, Yang Y, Wang J et al (2015) The CDKN2A-CDKN2B rs4977756 polymorphism and glioma risk: a meta-analysis. Int J Clin Exp Med 8(10):17480–17488
Mori K, Nakano M, Tokuda Y et al (2016) Stronger association of, CDKN2B-AS1, variants in female normal-tension glaucoma patients in a Japanese population. Invest Opthalmol Vis Sci 57(14):6416–6417. https://doi.org/10.1167/iovs.16-2041
Nunes HF, Ananina G, Costa VP et al (2017) Investigation of CAV1/CAV2 rs4236601 and CDKN2B-AS1 rs2157719 in primary open-angle glaucoma patients from Brazil. Ophthalmic Genet 39(2):1–6. https://doi.org/10.1080/13816810.2017.1393830
Vicente ZM, Carolina OA, Eva AM et al (2017) A multi-locus genetic risk score for primary open-angle glaucoma (POAG) variants is associated with POAG risk in a Mediterranean population: inverse correlations with plasma vitamin C and E concentrations. Int J Mol Sci 18(11):2302. https://doi.org/10.3390/ijms18112302
Funding
This work was supported by the National Natural Science Foundation of China (Grant 81271001).
Author information
Authors and Affiliations
Contributions
Data curation, methodology, software, validation: Liu SS and Chen SW. Project administration, Funding acquisition: Niu TT. Writing—original draft: Niu TT. Writing—review and editing: Liu SS and Chen SW.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, S., Chen, S. & Niu, T. Genetic association between CDKN2B-AS1 polymorphisms and the susceptibility of primary open-angle glaucoma (POAG): a meta-analysis from 21,775 subjects. Ir J Med Sci 191, 2385–2392 (2022). https://doi.org/10.1007/s11845-021-02794-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11845-021-02794-x