Abstract
COVID-19 has become a global pandemic caused by the SARS-CoV-2 coronavirus. SARS-CoV-2 shares many similarities with SARS coronavirus (SARS-CoV). A viral replication complex containing non-structural proteins (nsps) is the toolbox for RNA replication and transcription of both coronaviruses. In both cases, the RNA-dependent RNA polymerase (RdRp) domain of the coronaviral replication complex dictates the primary polymerase activity by cooperating with cofactors. The higher transmissibility and mortality due to SARS-CoV-2 are related to its higher RNA replication activity compared to SARS-CoV. The discrepancy between the RNA replication efficiency of SARS-CoV and SARS-CoV-2 can be understood by exploring interactions within their viral replication complexes. Our modeling of molecular interactions within the viral replication complexes of SARS-CoV and SARS-CoV-2 using molecular dynamics simulations suggests that in contrast to SARS-CoVnsp12, SARS-CoV2nsp12 prefers helices as the dominant interacting secondary motifs. The relative differences in nonbonded interactions between nsps could suggest viral RNA replication ability in coronaviruses.
Similar content being viewed by others
Introduction
From the beginning of the twenty-first century, the world has encountered several coronavirus infection outbreaks. During the last 20 years, three different coronavirus outbreaks resulting from zoonotic spillover (transmission from animal to humans) have caused considerable health-related problems, primarily by affecting the human respiratory system and leading to fatal outcome in some cases. SARS (Severe Acute Respiratory Syndrome), also named novel coronavirus SARS-CoV, emerged from China's Guangdong province in 2002–2003 and spread to 26 countries, infecting 8000 people, leading to the death of about 800 people.1 Another outbreak of zoonotic spillover coronavirus was marked by the emergence of MERS-CoV (Middle East Respiratory Syndrome) in Saudi Arabia in September 2012. The total number of reported cases of MERS-CoV was 2494, with 858 fatalities across 27 countries.2 Coronavirus Disease 2019 (COVID-19), caused by the novel coronavirus SARS-CoV-2, is the latest and most impactful addition to the list of zoonotic outbreaks. The first incidence of novel coronavirus SARS-CoV-2 was reported in December 2019 in Wuhan, China.3,4 Since then, this virus has infected people across 213 countries on all the continents except Antarctica. According to the most recent statistics (as of 18 January 2021) provided by the World Health Organization (WHO) COVID-19 dashboard, the total number of COVID-19 cases and deaths are 93,611,355 and 2,022,405, respectively.5 Although SARS-CoV and SARS-COV-2 coronaviruses have similarities in their origin and general structures and both target the respiratory system, there are dramatic differences in the infection rates and patient outcomes resulting from infections due to the two coronaviruses. The fatalities because of the SARS-COV-2 virus are 2,022,405 (as of 18 January 2021) and counting compared to the reported fatalities due to the SARS-COV of about 800.
Coronaviruses are enveloped, large, positive-stranded RNA viruses under the order of Nidovirales.6,7 This order contains four different virus families, i.e., Arteriviridae, Coronaviridae, Mesoniviridae, and Roniviridae. Coronavirinae and Torovirinae are the constituent sub-families of the Coronaviridae family.8,9 Alpha, beta, gamma, and delta coronaviruses are the four component genera of Coronavirinae.10,11 Coronaviruses accommodate the largest multicistronic RNA genome (27–32 kb) containing two open reading frames (ORF1a and ORF1b) at its 5′ side.12 Translation of ORF1a yields the replicative polyprotein 1a (pp1a), while the ribosomal frameshifting of ORF1a and ORF1b makes polyprotein 1b (pp1b).13,14 Co-translational proteolytic cleavage of polyproteins (pp1a and pp1ab) results in the formation of around 16 non-structural proteins (nsp1–16) that act as the viral replicating machinery.15,16 Structural proteins (spike, envelope, membrane, and nucleocapsid) of coronaviruses are predominantly encoded by the 3′ side of the RNA genome.17
The mechanism of host cell infection by viruses can be divided into five different stages. The very first stage is attachment, when a virus attaches to a host cell through a receptor protein. Then it enters into the host cell by either fusion or endocytosis. Following this, the viral proteins and genome are replicated and assembled inside the cell to make new viruses, followed by their release through lysis or exocytosis. Among these stages, viral replication is considered a critical stage for viral infection of tissues and organs inside the body. RNA-dependent RNA polymerase (RdRp), one of the most conserved elements of the coronaviral life cycle, has been considered the primary tool for the transcription and replication of the virus genome.18,19 RdRp encompasses about two-thirds of the non-structural protein 12 (nsp12).20 The length and location of nsp12 within the replicase polyprotein affect different features of coronaviruses.21 Two other non-structural proteins (nsp7 and nsp8) perform as the auxiliary components (cofactors) of the multi-subunit coronaviral replication complex led by nsp12.22 The participation of cofactors greatly enhances the polymerase activity of RdRp containing nsp12.23,24 Due to its RNA replication feature, RdRp has become the focus of pharmaceutical research as a potential drug target for coronavirus infections.25 For this reason, the use of remdesivir prodrug, as a suitable ligand of RdRp, has been of interest for SARS-CoV-2 treatment.24,26
SARS-CoV and SARS-CoV-2 diseases share many similarities as both the contributing coronaviruses are bat-originated beta-coronaviruses.27,28 Both of them follow the similar host cell attachment mechanism as their spike (S) protein binds to the cellular receptor angiotensin-converting enzyme 2 (ACE2).29,30 The nsp12–nsp7–nsp8 complex is utilized as the principal RNA replication/transcription toolbox by SARS-CoV and SARS-CoV-2, where nsp12 consists of RdRp.23,24 Though these two coronaviruses pursue almost identical viral lifecycles, the severity of SARS-CoV-2 is being observed to be enormously higher than that of SARS according to the number of reported cases and deaths. Moreover, these two coronaviruses have shown a marked difference in their replication efficiency, and it is still unclear what factors play decisive roles in this aspect.31 As the nsp12–nsp7–nsp8 complex is considered the hub of viral replication for both coronaviruses, the interactions within this viral protein complex need to be explored.
Remdesivir (GS-5734), an adenosine C-nucleoside prodrug, is considered a broad-spectrum antiviral agent against different RNA viruses.32 In 2017, Gilead Sciences developed the drug as a potential medication for Ebola viruses.33 This monophosphoramidate prodrug is effectively metabolized into active nucleoside triphosphate (GS-443902) when administered into human cell lines.34 This active ATP (adenosine triphosphate) analog causes the RNA chain termination by confusing viral RdRp and incorporating into the RNA chain.34,35 Remdesivir has attracted researchers' attention again because a recent in vitro study showed the inhibition of SARS-CoV-2, which utilized it as the inhibitor.26 Currently, remdesivir drug has been given emergency approval by the FDA in the USA to treat SARS-CoV-2 infected and hospitalized patients, as remdesivir-treated animals have shown lower lung damage than control animals36. As remdesivir is primarily targeted for the catalytic active site of nsp12 RdRp,37 binding of this prodrug can bring changes in the interactions within the viral complex. Hence, investigating the interactions within the protein subunits of the viral replication complex in the presence of remdesivir will help to explore its impact on the complex.
Molecular dynamics (MD) simulation is an in silico means of inspecting the structure and function of biomolecules.38,39,40,41,42,43 This is a computational technique that predicts the position and motion of atoms and molecules in a time-dependent manner utilizing Newton’s second law. MD simulation computes the molecular interactions through molecular mechanics that employs a force field containing energy constants.44 This study seeks to investigate the nonbonded (electrostatic and Van der Waals) molecular interactions among different subunits and domains inside the viral replication complex (nsp12–nsp7–nsp8) of SARS-CoV and SARS-CoV-2. This study also inspects these interactions when a remdesivir prodrug (remdesivir monophosphate/GS-441524 monophosphate) is in the proximity of RdRp domain of SARS-CoV-2 coronavirus.
Methodology
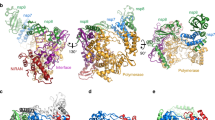
The initial three-dimensional models of SARS-CoV and SARS-CoV-2 viral replication complexes (nsp12–nsp7–nsp8 protein complex) have been obtained from the RCSB Protein Data Bank (PDB ID 6NUR23 and 6M7124, respectively). Each of the models contains one nsp12 polypeptide chain, one nsp7 chain, and two nsp8 chains. These analogous models are primarily chosen to compare the nonbonded interactions among the protein subunits of corresponding coronaviral replication complexes. The addition of hydrogen atoms and the assignment of partial charges were performed by the AutoPSF plugin of Visual Molecular Dynamics (VMD 1.9.4). Next, each of the models was solvated in a water box of 5 Å thickness on all sides to approximate the cellular environment. The dimensions of the solvated SARS-CoV and SARS-CoV-2 viral replication complexes are 87.15 Å × 91.74 Å × 126.52 Å and 83.81 Å × 95.22 Å × 117.82 Å, respectively (Fig. 1a and b). These solvated protein complex models were then simulated using NAMD 2.1245 from the Theoretical and Computational Biophysics Group developed NAMD at the Beckman Institute for Advanced Science and Technology at the University of Illinois at Urbana-Champaign. Initially, both models were minimized for achieving stable conformation using the conjugate gradient method.46 The solvated protein complexes were then brought to 310 K temperature and 1.01325 bar pressure to be consistent with the human physiological condition. The equilibrium condition was achieved at the same temperature and pressure by simulating both models for 5 ns with a timestep of 0.5 fs. Total energy denotes the thermodynamic equilibrium, while root-mean-square deviation (RMSD) specifies the conformational equilibrium. These equilibrated models were used to compute all the interactions among the subunits and domains within coronaviral replication complexes. CHARMM (Chemistry at HARvard Macromolecular Mechanics) force field was employed for computing all the interaction energies.47
Initial solvated models of viral replication complexes (nsp12–nsp7–nsp8) of (a) SARS-CoV (6NUR); (b) SARS-CoV-2 (6M71); (c) SARS-CoV-2 without remdesivir monophosphate (RMP) (7BV2); (d) SARS-CoV-2 with RMP (7BV2) as well as highlighted RMP. In all figures, the blue, red, and pink colored chains represent the full-length nsp12, nsp8, and nsp7, respectively. The green-colored chain represents the second monomer of nsp8 for the first two models (6NUR and 6M71).
To probe the contribution of remdesivir prodrug on the interactions within the SARS-CoV-2 viral replication complex, a different set of structures have been used. The apo (ligand-free) form (PDB ID 7BV137) and ligand-bound form (PDB ID 7BV237) of SARS-CoV-2 viral complexes are obtained from Protein Data Bank. However, these structures are not analogous as the apo form (7BV1) contains two chains of nsp8 while the ligand-bound form (7BV2) consists of only one chain of nsp8. As the overall structure of apo viral complex (7BV1) is similar to the ligand bound structure (7BV2),37 we only utilize the latter model in both the ligand-bound and ligand-removed conditions. The protein interactions within the ligand-bound replication complex are compared with those of the ligand-removed complex. Both protein complexes (ligand-bound and ligand-removed) are assigned hydrogens and partial charges, solvated in 5 Å thick water boxes, minimized, and later equilibrated for 5 ns at the physiological conditions (310 K and 1.01325 bar) utilizing NAMD and CHARMM force field. The initial dimensions of the solvated ligand-bound and ligand-removed complexes are essentially the same (87.75 Å × 92.64 Å × 106.08 Å) as the ligand stays in the cavity of nsp12 (Fig. 1c and d). All the solvated models consist of TIP3P water molecules.
Although an ideal ligand should be an active metabolite of remdesivir (remdesivir triphosphate/GS-443902), remdesivir monophosphate (RMP)/GS-441524 monophosphate has been chosen as the ligand in the present study as it is the only available ligand model experimentally docked into SARS-CoV-2 viral replication complex37 and deposited into Protein Data Bank as PDB ID 7BV2. Therefore, we will be performing MD simulations on one model of 7BV2 without RMP (Non-RMP model) and another model of 7BV2 with RMP (RMP bound model). RMP (C12H14N5O7P) is a small organic molecule with a molecular weight of 371.24 amu. The parameterization of RMP is performed by the CHARMM General Force Field (CGenFF) program and the Force Field Toolkit (ffTK) plugin of visual molecular dynamics (VMD). Initially, the atom types are determined by CGenFF program,47 which is rule-based and programmable. Next, utilizing the ffTK48 plugin of VMD 1.9.4 and CGenFF topology and parameters, all the atoms are assigned Lennard–Jones parameters according to their atom types. Using the same methodology, the non-polar hydrogen atoms are assigned a partial charge of + 0.09 while all the other atoms are initially assigned a 0.00 charge. Following this step, geometry optimization (quantum mechanical calculations) of RMP is performed using the Gausssian09 software package to obtain the lowest energy conformation. This optimized geometry is utilized to derive partial atomic charges of the remaining atoms by reproducing the explicit interactions with TIP3P water molecules. These quantum mechanical (QM) water interaction energies of RMP are used to obtain the optimized CHARMM partial charges. The bond stretching and angle bending potentials are derived by performing QM Hessian calculations which employ second derivative of potential energy to model the potential energy surfaces representing the bond and angle distortions. The dihedral parameters of RMP are obtained by QM scanning of the torsional potential energy surfaces. The water interaction energy calculations utilize the Hartree–Fock (HF) theory, while all the bonded parameter potential energy calculations utilize the second-order Møller–Plesset perturbation theory (MP2). All the QM calculation inputs are generated by ffTK plugin, and the simulations are conducted using Gaussian09 software. The calculated CHARMM force field parameters for remdesivir monophosphate (RMP) are presented in Supplementary Tables S-XV to S-XVIX.
We have used the Center for Computationally Assisted Science and Technology (CCAST), a supercomputing cluster at North Dakota State University, to perform all the MD and QM simulations. Each of the simulations was run on 1 node and 20 Intel Xeon 2.5 GHz processors with 20 GB DDR3 RAM at 1866 MHz.
Results
SARS-CoV Replication Complex
The equilibrium structure of the SARS-CoV viral replication complex consists of one nsp12 chain (A), one nsp7 chain (C), and two nsp8 chains (B, D) (Fig. 2).23 The polypeptide chain of nsp12 (A) is 793 amino acids long as obtained from the Protein Data Bank (6NUR). The length of the nsp7 chain (C) and np8 chain (B) is 70 and 115 residues, respectively. The second subunit of nsp8 (chain D) is 109 residues long. The structure of nsp12 can be divided into three different domains, i.e., nucleotidyltransferase (NiRAN), interface, and RdRp domain. The nidovirus RdRp-associated NiRAN domain is positioned at the N-terminal of nsp12 (residues 117-250) while the RNA-dependent RNA polymerase (RdRp) domain lies at its C-terminal (residues 399–920). The interface domain (residues 251–398) is the connecting domain between the NiRAN and RdRp domain. RdRp domain performs the primary polymerase activity of SARS-CoV coronavirus with the assistance of cofactors.23 The nonbonded interactions (electrostatic and Van der Waals) have been computed within this SARS-CoV viral replication complex to better understand the relationship among different nsps and domains.
Equilibrated structure of SARS-CoV (6NUR) viral replication complex (nsp12–nsp7–nsp8). The purple, cyan, and blue colored segments represent the NiRAN, Interface, and RNA-dependent RNA polymerase (RdRp) domains of nsp12 (chain A). The red and green chains (chain B and D, respectively) represent two monomers of nsp8. The nsp7 is represented by a pink-colored chain (chain C). For the convenience of visualization, the water molecules are not shown.
The total nonbonded interactions between nsp12 and its cofactors (nsp7 and nsp8) are − 812.8 kcal/mol where electrostatic, and Van der Waals (VDW/VdW) interactions are − 509.1 kcal/mol and − 303.7 kcal/mol, respectively (Supplementary Table S-I). Three different residues (GLU, ASP, LEU) of nsp12 contribute to more than half of these interaction energies (53.1%) by interacting with the cofactors. The amounts of non-bonded interaction energies contributed by GLU, ASP and LEU of nsp12 are − 209.9 kcal/mol (25.8%), − 122.3 kcal/mol (15.1%), and − 105.2 kcal/mol (12.9%), respectively. PHE, ASN, ARG, PRO, and TYR are other significantly interacting residues, i.e., each of them contributed to more than 4% of the total interactions (Fig. 3a and d). In terms of the nsp12 secondary structure, the sheets interact with the cofactors most (− 266.6 kcal/mol) while coils interact the least (− 145 kcal/mol) (Fig. 3b and e). About two-thirds of all the interaction energies (− 530.4 kcal/mol) result from the polar residues of nsp12, while the rest (− 282.4 kcal/mol) are caused by the hydrophobic residues (Fig. 3c and f).
Non-bonded interaction (electrostatic and Van der Waals) energies of nsp12 with the co-factors (nsp7 and nsp8) in terms of its (a) primary, (b) secondary, and (c) tertiary motifs within the equilibrated SARS-CoV viral replication complex (6NUR). Relative contribution (proportion) of interactions made by each (d) primary, (e) secondary, and (f) tertiary motif of nsp12. (g) Total non-bonded interactions of nsp12 with nsp7 and nsp8; (h) domain-wise interactions of nsp12 with nsp7; (i) domain-wise interactions of nsp12 with nsp8.
The total non-bonded interactions of nsp12 with its co-factors (− 812.8 kcal/mol) are divided into nsp12–nsp7 interactions (− 303.5 kcal/mol) and nsp12–nsp8 interactions (− 509.3 kcal/mol) (Fig. 3g). The total interaction energies between nsp7 and nsp8 are found to be − 436.9 kcal/mol. The total interaction energies of nsp12 with cofactors are also analyzed in terms of the three domains of nsp12. The NiRAN domain does not interact with the cofactors (Fig. 3h and i). The interface domain of nsp12 contributes to 43% (− 349.5 kcal/mol) while RdRp domain contributes to 57% (− 463.3 kcal/mol) of total nsp12-cofactor interaction energies. The interface domain of nsp12 does not interact with nsp7 (Fig. 3h). The interaction energies of the RdRp domain of nsp12 with nsp7 and nsp8 have been calculated as − 303 kcal/mol and − 160.3 kcal/mol, respectively.
SARS-CoV-2 Replication Complex
The replication complex of SARS-CoV-2 has a similar structure to SARS-CoV. The equilibrated SARS-CoV-2 replication complex contains one chain of nsp12 (A), one chain of nsp7 (C), and two chains of nsp8 (B&D).24 The nsp12 chain (A) consists of 851 residues, while the nsp7 chain (C) contains 70 residues. Chain B of nsp8 has 113 residues. The single monomer of nsp8 (chain D) comprises of only 43 residues. NiRAN (residues 60–249), interface (residues 250–365), and RdRp (residues 366–920) domains create the nsp12 structure (Fig. 4). The interface domain serves as a connecting domain between NiRAN and RdRp. Like SARS-CoV, the RdRp domain of SARS-CoV-2 is primarily responsible for viral replication in the presence of cofactors.24
Equilibrated structure of SARS-CoV-2 (6M71) viral replication complex (nsp12–nsp7–nsp8). The purple, cyan, and blue colored segments represent the NiRAN, interface, and RNA-dependent RNA polymerase (RdRp) domains of nsp12 (chain A). The red and green chains (chain B and D, respectively) represent two monomers of nsp8. The nsp7 is represented by a pink-colored chain (chain C). For the convenience of visualization, the water molecules are not shown.
The nsp12 monomer interacts with cofactors (nsp7 and nsp8) with nonbonded energy of − 672 kcal/mol (Supplementary Table S-IV). Electrostatic interactions contributed 57% (− 384 kcal/mol) of the interaction energies while VDW furnished the rest (− 288 kcal/mol). ASP (− 195.5 kcal/mol), LEU (− 105.3 kcal/mol), and GLU (− 66.8 kcal/mol) residues of nsp12 together result in about 54.7% of the interaction energies. ALA, ASN, LYS, PHE, PRO, and VAL residues of nsp12 individually provide at least 4% of the interaction energies (Fig. 5a and d). The helices of nsp12 interact with cofactors with an energy of − 223.2 kcal/mol, which is 33.2% of the total interactions. The sheets of nsp12 interact in almost the same order as that of helices (− 211.7 kcal/mol) followed by coils (− 138.2 kcal/mol) (Fig. 5b and e). Polar residues of nsp12 yield approximately 59% (− 396.4 kcal/mol) of the interaction energies (Fig. 5c and f).
Nonbonded interaction energies (electrostatic and Van der Waals) of nsp12 with the cofactors (nsp7 and nsp8) in terms of its (a) primary, (b) secondary, and (c) tertiary motifs within the equilibrated SARS-CoV-2 viral replication complex (6M71). Relative contribution (proportion) of interactions made by each (d) primary, (e) secondary, and (f) tertiary motif of nsp12. (g) Total non-bonded interactions of nsp12 with nsp7 and nsp8; (h) domain-wise interactions of nsp12 with nsp7; (i) domain-wise interactions of nsp12 with nsp8.
The magnitudes of nonbonded interactions between nsp12–nsp7 (A-C) and nsp12–nsp8 (A-BD) have been computed as − 164.7 kcal/mol and − 507.3 kcal/mol, respectively (Fig. 5g). The nonbonded interactions between nsp7 and nsp8 are computed as − 476.9 kcal/mol. The NiRAN domain interacts with neither of the cofactors, while the interface domain interacts with only nsp8 (− 186.1 kcal/mol) (Fig. 5h and i). The RdRp domain of nsp12 yields 72.3% (− 485.9 kcal/mol) of its total interactions with the cofactors. The interactions of RdRp with nsp7 and nsp8 have been calculated as − 164.7 kcal/mol and − 321.2 kcal/mol, respectively.
Non-RMP and RMP-Bound SARS-CoV-2 Replication Complex
This viral replication complex of SARS-CoV-2 consists of one nsp12 chain (A), one nsp8 chain (B), and one nsp7 chain (C) (7BV2). The nsp12 chain is composed of NIRAN (residues 115–250), interface (residues 251–365), and RdRp (residues 366–920) domains.37 Remdesivir monophosphate (RMP), the potent antiviral prodrug, is removed from the ‘Non-RMP’ model of replication complex while kept in the native conformation in the ‘RMP-bound’ model of replication complex. The CHARMM compatible parameters of RMP, obtained through rigorous QM calculations and multidimensional optimization, are provided in the Supplementary Materials (refer to online supplementary material, Supplementary Table S-XV to S-XIX).
In the equilibrated ‘Non-RMP’ model (Fig. 6a), the total interaction energy of nsp12 with its cofactors (nsp7 and nsp8) is found to be − 929 kcal/mol. Almost 80% of the interaction energies (− 742.2 kcal/mol) are contributed by nsp12–nsp8 interactions while nsp12–nsp7 generates the rest (− 186.8 kcal/mol) (Fig. 6c). The NiRAN domain of nsp12 has no interactions with cofactors, while the interface domain only interacts with nsp8 (− 317 kcal/mol) (Fig. 6d). The RdRp domain is responsible for all the interactions of nsp12 with nsp7 (− 186.8 kcal/mol), while this domain provides a relatively higher portion (58%) of interactions (− 425.2 kcal/mol) between nsp12 and nsp8 compared to the interface domain (42%). The interactions between nsp7 and nsp8 are computed as − 87.8 kcal/mol.
Equilibrated structures of (a) non-RMP and (b) RMP-bound SARS-CoV-2 (7BV2) viral replication complexes (nsp12–nsp7–nsp8). The purple, cyan, and blue colored segments represent the NiRAN, interface, and RNA-dependent RNA polymerase (RdRp) domains of nsp12 (chain A). The red and pink chains (chain B and C) represent nsp8 and nsp7, respectively. The small licorice structure is identified as RMP in the ‘RMP-bound’ model. For the convenience of visualization, the water molecules are not shown. (c) Total nonbonded interactions of nsp12 with nsp7 and nsp8 in the ‘Non-RMP’ model. (d) Domain-wise nonbonded interactions of nsp12 with nsp7 and nsp8 in the ‘Non-RMP’ model. (e) Total nonbonded interactions of nsp12 with nsp7 and nsp8 in the ‘RMP-bound’ model. (f) Domain-wise nonbonded interactions of nsp12 with nsp7 and nsp8 in the ‘RMP-bound’ model.
In the equilibrated ‘RMP-bound’ model (Fig. 6b), the antiviral ligand RMP stays in the proximity of the catalytic active site of nsp12 RdRp. In this model, the interactions of nsp12–nsp8 and nsp12–nsp7 are determined as − 566.7 kcal/mol and − 298.6 kcal/mol, respectively (Fig. 6e), leading to a total nsp12-cofactor interaction of − 865.3 kcal/mol. The nsp12–nsp7 interactions are fully contributed by RdRp while nsp12–nsp8 interactions are contributed by both interface (− 188.8 kcal/mol) and RdRp (− 377.9 kcal/mol) domains (Fig. 6f). The nsp7–nsp8 interactions are determined as − 85.5 kcal/mol. The antiviral prodrug RMP is only observed to have interactions with nsp12 RdRp. The total nonbonded interaction energy between the RdRp domain and RMP is determined as − 206.9 kcal/mol, 95% of which results from electrostatic interactions.
Discussion
Protein-protein interactions are at the core of most physiological processes in the biological realm.49 Viral protein-protein interactions dictate different host cell infection stages, i.e., replication and assembly of virions and their resistance against the host immune system.50 These protein–protein interactions are divided into covalent (bonded) and non-covalent (nonbonded) interactions at the molecular level. Non-covalent (nonbonded) interactions play critical roles in genome replication, protein folding, signal transduction, enzyme detection, and many other physicochemical processes.51,52,53 Therefore, investigating the nonbonded viral protein–protein interactions in coronaviruses will help better perceive the biological activities (i.e., replication) carried out by the viral replication protein complexes.
SARS-CoV and SARS-CoV-2 coronaviruses comprise the analogous structure of the viral replication complex (nsp12–nsp7–nsp8). In both cases of coronaviruses, a peptide chain of nsp12 (A) performs as the principal replication subunit while one chain of nsp7 (C) and two chains of nsp8 (B, D) act as the replication cofactors.23,24 The structural conformation of cofactors can be described as a hetero-dimer of nsp7–nsp8 (C-B) along with an nsp8 monomer (D) for both coronaviral replication complexes. Significant attractive nonbonded (electrostatic and VDW) interactions are observed among the subunits of the replication complex. The nonbonded interactions of SARS-CoV nsp12 with its cofactors (− 812.8 kcal/mol) are higher than the interactions of SARS-CoV-2 nsp12 with its cofactors (− 672 kcal/mol). Electrostatic interactions yield a greater portion of the nonbonded interactions for both SARS-CoV (62.6%) and SARS-CoV-2 (57.1%). The peptide chain of nsp12 accomplishes more than half of its interactions with cofactors through three different residues, i.e., ASP, GLU, and LEU. GLU is the highest interacting (− 209.9 kcal/mol) residue of SARS-CoV nsp12, and ASP is the highest interacting residue (− 195.5 kcal/mol) of SARS-CoV-2 nsp12. CYS has been found as the least interacting residue of nsp12 for both coronaviruses. Regarding the secondary structure of nsp12, the highest portion of its interactions is undertaken by beta sheets (− 266.6 kcal/mol) and helices (− 223.2 kcal/mol) for SARS-CoV and SARS-CoV-2, respectively. The relative contribution by nsp12 sheets remained identical (~ 32%) for both coronaviruses. Polar residues of SARS-CoV nsp12 are attributed to relatively higher interactions (65.3%) than those of SARS-CoV-2 nsp12 (59%).
The interactions of nsp12 with cofactors are analyzed to have a better understanding of the viral replication complexes. The interactions between nsp12 (A) and nsp7 (C) are significantly higher for SARS-CoV (− 303.5 kcal/mol) compared to SARS-CoV-2 (− 164.7 kcal/mol) (Fig. 7a). The SARS-CoV nsp12 interacts with two nsp8 chains (B, D) almost as equally as SARS-CoV-2, i.e., the nsp12–nsp8 interactions are − 509.3 kcal/mol and − 507.3 kcal/mol for SARS-CoV and SARS-CoV-2, respectively. There are both similarities and differences between the coronaviruses in terms of their domain-wise nsp12 interactions with cofactors. The NiRAN domain does not interact with cofactors in the replication complex of both coronaviruses. The majority of coronavirus nsp12 interactions (with cofactors) are performed by its RdRp domain. The RdRp domain of nsp12 carries out the primary polymerase activity in the presence of cofactors. The RdRp domain of SARS-CoV-2 exhibits similar interactions (− 485.9 kcal/mol) with the two cofactors compared to the SARS-CoV RdRp (− 463.3 kcal/mol). However, the proportion of RdRp domain interactions with cofactors towards the total nsp12 interactions is significantly higher for SARS-CoV-2 (72.3%) than SARS-CoV (57%). The interactions of SARS-CoV-2 RdRp with nsp7 co-factor (− 164.7 kcal/mol) are 45.6% lower than that of SARS-CoV (− 303.5 kcal/mol). On the contrary, the interactions of SARS-CoV-2 RdRp with nsp8 cofactor (− 321.2 kcal/mol) are approximately 100% higher than those of SARS-CoV (− 160.3 kcal/mol). Thus, significant differences are observed in the nsp12 RdRp domain interactions with the cofactors nsp7 and nsp8 between SARS-COV and SARS-COV-2.
Comparison of domain-wise interaction energies of nsp12 with cofactors (nsp7 and nsp8) between equilibrated (a) SARS-CoV (6NUR) and SARS-CoV-2 (6M71) viral replication complexes and (b) non-RMP (without RMP) and RMP-bound (with RMP) SARS-CoV-2 (7BV2) viral replication complexes. All energies have the unit of kcal/mol.
The replication and transcription procedure of the coronavirus RNA genome is guided by the replication complex interactions (nsp12–nsp7–nsp8).22,23 Therefore, the severity of coronavirus infections can result from a higher replication/transcription rate inside the host cells. SARS-CoV and SARS-CoV-2 coronaviruses have shown different replication trends in different human cells. SARS-CoV exhibits higher replication in intestinal CaCo2 cells, while SARS-CoV-2 demonstrates higher virus production in pulmonary Calu3 cells.54 SARS-CoV-2 has also been reported to replicate more extensively in bronchial epithelium compared to SARS-CoV. This particular observation has been considered as one of the reasons for the higher transmissibility of SARS-CoV-2.55 Although it is known that the interactions between the non-structural protein complex (nsp12–nsp7–nsp8) are key to the replication of viral RNA in cells, little is known about how the RdRp domain interactions specifically with nsp7 or nsp8 contribute to replication in different types of cells. It might be possible that higher RdRp–nsp7 interactions can contribute to the higher replication of SARS-CoV RNA in specific organs or cell lines. On the other hand, higher replication of SARS-CoV-2 RNA in certain parts of the body can be related to higher RdRp–nsp8 interactions. The results from this work suggest investigating the replication rates and their relationship to RdRp domain interactions with nsp7 and nsp8 for different cell types through carefully crafted experiments. It is important to note that the differences in amino acid compositions of nsp12 between SARS-CoV and SARS-CoV-2 do not contribute to their interactions as they share about 96% sequence identity, and the different residues mostly lie in distant regions from the cofactors (supplementary Fig. S-2). The variations in the non-structural proteins' interactions may be attributed to their conformation and relative orientations within the viral replication complex.
Comparison between the ‘Non-RMP’ and ‘RMP-bound’ SARS-CoV-2 viral replication complexes (7BV2) indicates the impact of antiviral prodrug RMP on interaction energies inside the RdRp complex. The presence of RMP reduces the interaction energy between nsp12 and nsp8 from − 742.2 kcal/mol to − 566.7 kcal/mol (Fig. 7b). This reduction in interaction energy can also be seen within the interacting individual domains of nsp12. The RdRp domain of nsp12 interacts less (− 377.9 kcal/mol) with nsp8 in the RMP-bound model compared to the Non-RMP model (− 425.2 kcal/mol). The study results comparing SARS-CoV and SARS-CoV-2 presented earlier have indicated that lower RdRp domain–nsp8 interactions in SARS-CoV versus SARS-CoV-2 may have contributed to the lower replication of RNA in SARS-CoV compared to SARS-CoV-2. Based on this co-relationship, the reduction of RdRp–nsp8 interactions in the presence of RMP can reduce coronaviral replication in case of SARS-CoV-2 infection. The presence of RMP also decreases the interactions between the nsp8 and interface domain significantly while increasing the interactions between RdRp and nsp7. In a recent study, remdesivir has been shown to inhibit SARS-CoV-2 replication in human lung cells and the primary epithelial airways.56 The primary inhibition mechanism of active remdesivir metabolite (remdesivir triphosphate/RTP) is to terminate RNA chain synthesis by interfering with the nsp12 polymerase (RdRp) activity.57 The presence of RMP ligand in the catalytic active site of RdRp modifies its interactions with cofactors, which may lead to the disruption of its regular polymerase activity.
In summary, SARS-CoV nsp12 has higher non-bonded interaction energies (− 812 kcal/mol) with co-factors (nsp7 and nsp8) compared to SARS-CoV-2 (− 672 kcal/mol). The coronaviral nsp12 consists of three different domains: nucleotidyltransferase (NiRAN), interface, and RNA-dependent RNA polymerase (RdRp) domain. Among these three domains, only the RdRp domain is attributed to viral replication.23,24 Therefore, only the interactions between the RdRp domain of nsp12 and co-factors need to be considered for viral RNA replication. SARS-CoV RdRp has lower interactions (− 463.3 kcal/mol) with co-factors compared to SARS-CoV-2 (− 485.9 kcal/mol). This finding is consistent with higher infectious behavior of SARS-CoV-2. Moreover, by analyzing the pairwise interactions of RdRp with individual co-factor nsp8, it has been observed that SARS-CoV-2 RdRp–nsp8 interaction (− 321.2 kcal/mol) is almost twice of that of SARS-CoV (− 160.3 kcal/mol). This specific interaction (RdRp–nsp8) might play a key role in higher replication of SARS-CoV-2 in bronchial epithelium, which in turn may lead to higher transmissibility of SARS-CoV-2.55 The presence of RMP within SARS-CoV-2 viral replication complex has been observed to reduce the interactions between RdRp and nsp8. This reduction in interactions can contribute to the reduction of viral RNA replication activity.
Conclusion
The invasion of new host cells in coronavirus infections depends on the viral replication complex's virus reproduction process. This coronaviral replication complex comprises three non-structural proteins (nsp12–nsp7–nsp8) that replicate and transcribe new RNA genomes. For the first time, we have compared differences in interactions of viral replication complexes in SARS-CoV and SARS-CoV-2 using molecular dynamics simulations. As remdesivir prodrug is primarily targeted to the catalytic active site of nsp12 RdRp, remdesivir prodrug (RMP) treated animals have shown lower lung damage compared to control animals. Here we have also evaluated the impact of remdesivir prodrug (RMP) on the SARS-CoV-2 viral complex to find the differences between the interactions within the nsps with and without RMP. The significant findings of this study are listed below:
-
1.
In both viral replication complexes, the peptide chain of nsp12 contributes to more than half of its interactions with cofactors (nsp7 and nsp8) through ASP, GLU, and LEU residues.
-
2.
Sheets are the most interacting secondary motifs of SARS-CoV nsp12, while in SARS-CoV-2, helices contribute to the highest interactions.
-
3.
The interaction energies between the nsps in the viral replication complex are significantly different, potentially a clue for the differences seen in the RNA replication rates exhibited by the two coronaviruses. The RdRp–nsp7 interaction energies of SARS-CoV are twice those of SARS-CoV-2. On the other hand, the RdRp–nsp8 interaction energies of SARS-CoV are half of those of SARS-CoV-2.
-
4.
Remdesivir monophosphate (RMP) reduces the RdRp–nsp8 interactions of the SARS-CoV-2 viral complex. Reduced RdRp–nsp8 interactions may contribute to reduced RNA replication rates in SARS-CoV-2, consequently resulting in reduced infections and mortality.
-
5.
This work points towards the potential for using the relative differences in nonbonded interactions between nsps as an indicator of viral RNA replication ability in coronavirus infections. Well-designed experiments based on the simulation results would provide further credence to this assertion.
References
M. Chan-Yeung and R.H. Xu, Respirology 8(Suppl S9), 14. (2003).
J. Lee, G. Chowell, and E. Jung, J. Theor. Biol. 408, 118. (2016).
W. Tan, X. Zhao, X. Ma, W. Wang, P. Niu, W. Xu, G.F. Gao, and G.Z. Wu, China CDC Wkly. 2(4), 61. (2020).
N. Zhu, D. Zhang, W. Wang, X. Li, B. Yang, J. Song, X. Zhao, B. Huang, W. Shi, R. Lu, P. Niu, F. Zhan, X. Ma, D. Wang, W. Xu, G. Wu, G.F. Gao, and W. Tan, N. Engl. J. Med. 382(8), 727. (2020).
W.H. Organization, WHO Coronavirus (COVID-19) Dashboard, 2021. https://covid19.who.int/. Accessed 18 January 2021.
M.M.C. Lai and K.V. Holmes, in Fields Virology. ed. by D.M. Knipe, and P.M. Howley (Lippincott Williams & Wilkins, Philadelphia, 2001), pp. 1163–1179.
J.M. Gonzaalez, P. Gomez-Puertas, D. Cavanagh, A.E. Gorbalenya, and L. Enjuanes, Arch. Virol. 148(11), 2207. (2003).
R. Draker, R.L. Roper, M. Petric, and R. Tellier, Virus Res. 115(1), 56. (2006).
S.Z.J. Siddell and E.J. Snijder, Coronaviruses, Toroviruses, and Arteriviruses, vol 1. (Hodder Arnold, London, 2005).
P.C.Y. Woo, S.K.P. Lau, C.S.F. Lam, C.C.Y. Lau, A.K.L. Tsang, J.H.N. Lau, R. Bai, J.L.L. Teng, C.C.C. Tsang, and M. Wang, J. Virol. 86(7), 3995. (2012).
G. Li, Y. Fan, Y. Lai, T. Han, Z. Li, P. Zhou, P. Pan, W. Wang, D. Hu, and X. Liu, J. Med. Virol. 92(4), 424. (2020).
P.T. Nga, M. del Carmen Parquet, C. Lauber, M. Parida, T. Nabeshima, F. Yu, N.T. Thuy, S. Inoue, T. Ito, and K. Okamoto, PLoS Pathog. 7(9), e1002215. (2011).
P.S. Masters, Adv. Virus Res. 66, 193. (2006).
A.E. Firth and I. Brierley, J. Gen. Virol. 93(Pt 7), 1385. (2012).
E.J. Snijder, P.J. Bredenbeek, J.C. Dobbe, V. Thiel, J. Ziebuhr, L.L.M. Poon, Y. Guan, M. Rozanov, W.J.M. Spaan, and A.E. Gorbalenya, J. Mol. Biol. 331(5), 991. (2003).
V. Thiel, K.A. Ivanov, A. Putics, T. Hertzig, B. Schelle, S. Bayer, B. Weißbrich, E.J. Snijder, H. Rabenau, and H.W. Doerr, J. Gen. Virol. 84(9), 2305. (2003).
A.M.Q. King, E. Lefkowitz, M.J. Adams, and E.B. Carstens, Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses, vol 9. (Elsevier, Amsterdam, 2011).
D.-G. Ahn, J.-K. Choi, D.R. Taylor, and J.-W. Oh, Arch. Virol. 157(11), 2095. (2012).
S.M. Brockway, C.T. Clay, X.T. Lu, and M.R. Denison, J. Virol. 77(19), 10515. (2003).
A. Velthuis, J.J. Arnold, C.E. Cameron, S.H.E. van den Worm, and E.J. Snijder, Nucleic Acids Res. 38(1), 203. (2010).
E.J. Snijder, E. Decroly, and J. Ziebuhr, Adv. Virus Res. 9, 59. (2016).
L. Subissi, C.C. Posthuma, A. Collet, J.C. Zevenhoven-Dobbe, A.E. Gorbalenya, E. Decroly, E.J. Snijder, B. Canard, and I. Imbert, Proc. Natl. Acad. Sci. U.S.A. 111(37), E3900. (2014).
R.N. Kirchdoerfer and A.B. Ward, Nat. Commun. 10(1), 1. (2019).
Y. Gao, L.M. Yan, Y.C. Huang, F.J. Liu, Y. Zhao, L. Cao, T. Wang, Q.Q. Sun, Z.H. Ming, L.Q. Zhang, and J. Ge, Science 368(6492), 779. (2020).
W.-J. Guan, Z.-Y. Ni, Y. Hu, W.-H. Liang, C.-Q. Ou, J.-X. He, L. Liu, H. Shan, C.-L. Lei, and D.S.C. Hui, N. Engl. J. Med. 382(18), 1708. (2020).
M. Wang, R. Cao, and L. Zhang, Cell Res. 30, 269–271. (2020).
P. Zhou, X.-L. Yang, X.-G. Wang, B. Hu, L. Zhang, W. Zhang, H.-R. Si, Y. Zhu, B. Li, C.-L. Huang, H.-D. Chen, J. Chen, Y. Luo, H. Guo, R.-D. Jiang, M.-Q. Liu, Y. Chen, X.-R. Shen, X. Wang, X.-S. Zheng, K. Zhao, Q.-J. Chen, F. Deng, L.-L. Liu, B. Yan, F.-X. Zhan, Y.-Y. Wang, G.-F. Xiao, and Z.-L. Shi, Nature 579, 270. (2020).
B. Hu, L.-P. Zeng, X.-L. Yang, X.-Y. Ge, W. Zhang, B. Li, J.-Z. Xie, X.-R. Shen, Y.-Z. Zhang, N. Wang, D.-S. Luo, X.-S. Zheng, M.-N. Wang, P. Daszak, L.-F. Wang, J. Cui, and Z.-L. Shi, PLoS Pathog. 13(11), e1006698. (2017).
W.H. Li, M.J. Moore, N. Vasilieva, J.H. Sui, S.K. Wong, M.A. Berne, M. Somasundaran, J.L. Sullivan, K. Luzuriaga, T.C. Greenough, H. Choe, and M. Farzan, Nature 426(6965), 450. (2003).
J. Lan, J. Ge, J. Yu, S. Shan, H. Zhou, S. Fan, Q. Zhang, X. Shi, Q. Wang, and L. Zhang, Nature 581(7807), 215. (2020).
P. V’kovski, A. Kratzel, S. Steiner, H. Stalder, and V. Thiel, Nat. Rev. Microbiol. 19, 1. (2020).
D. Siegel, H.C. Hui, E. Doerffler, M.O. Clarke, K. Chun, L. Zhang, S. Neville, E. Carra, W. Lew, and B. Ross, J. Med. Chem. 60(5), 1648. (2017).
J.A. Al-Tawfiq, A.H. Al-Homoud, and Z.A. Memish, Travel Med. Infect. Dis. 34, 10615. (2020).
T.K. Warren, R. Jordan, M.K. Lo, A.S. Ray, R.L. Mackman, V. Soloveva, D. Siegel, M. Perron, R. Bannister, and H.C. Hui, Nature 531(7594), 381. (2016).
E.P. Tchesnokov, J.Y. Feng, D.P. Porter, and M. Götte, Viruses 11(4), 326. (2019).
B.N. Williamson, F. Feldmann, B. Schwarz, K. Meade-White, D.P. Porter, J. Schulz, N. van Doremalen, I. Leighton, C.K. Yinda, L. Pérez-Pérez, A. Okumura, J. Lovaglio, P.W. Hanley, G. Saturday, C.M. Bosio, S. Anzick, K. Barbian, T. Cihlar, C. Martens, D.P. Scott, V.J. Munster, and E. de Wit, Nature 585(7824), 273. (2020).
W. Yin, C. Mao, X. Luan, D.-D. Shen, Q. Shen, H. Su, X. Wang, F. Zhou, W. Zhao, and M. Gao, Science 368(6498), 1499. (2020).
C. Sagui, and T.A. Darden, Annu. Rev. Biophys. Biomol. Struct. 28(1), 155. (1999).
M. Karplus, and J.A. McCammon, Nat. Struct. Biol. 9(9), 646. (2002).
A. Sharma, M.L. Snead, K.S. Katti, and D.R. Katti, Chem. Phys. 522, 104. (2019).
S.M. Pradhan, K.S. Katti, and D.R. Katti, Biomacromol 13(8), 2562. (2012).
P. Ghosh, D.R. Katti, and K.S. Katti, Biomacromol 8(3), 851. (2007).
S.V. Jaswandkar, H.M.N. Faisal, K.S. Katti, and D.R. Katti, Biomacromol 22, 902. (2021).
D. Young, Computational Chemistry: A Practical Guide for Applying Techniques to Real World Problems (Wiley, Hoboken, 2004).
J.C. Phillips, R. Braun, W. Wang, J. Gumbart, E. Tajkhorshid, E. Villa, C. Chipot, R.D. Skeel, L. Kale, and K. Schulten, J. Comput. Chem. 26(16), 1781. (2005).
M.C. Payne, M.P. Teter, D.C. Allan, T.A. Arias, and J.D. Joannopoulos, Rev. Mod. Phys. 64(4), 1045. (1992).
K. Vanommeslaeghe, E. Hatcher, C. Acharya, S. Kundu, S. Zhong, J. Shim, E. Darian, O. Guvench, P. Lopes, I. Vorobyov, and A.D. MacKerell, J. Comput. Chem. 31(4), 671. (2010).
C.G. Mayne, J. Saam, K. Schulten, E. Tajkhorshid, and J.C. Gumbart, J. Comput. Chem. 34(32), 2757. (2013).
A.C. Pan, D. Jacobson, K. Yatsenko, D. Sritharan, T.M. Weinreich, and D.E. Shaw, Proc. Natl. Acad. Sci. U.S.A. 116(10), 4244. (2019).
J.A. Pan, X.X. Peng, Y.J. Gao, Z.L. Li, X.L. Lu, Y.Z. Chen, M. Ishaq, D. Liu, M.L. De Diego, L. Enjuanes, and D.Y. Guo, PLoS ONE 3(10), e3299. (2008).
K.E. Riley, and P. Hobza, Wiley Interdiscip. Rev. Comput. Mol. Sci. 1(1), 3–17. (2011).
A. Erijman, E. Rosenthal, and J.M. Shifman, PLoS ONE 9(10), e110085. (2014).
L. Stryer, J.L. Tymoczko, and J.M. Berg, Biochemistry, 5th edn. (Freeman, WH and Company, New York, 2002), p 41.
H. Chu, J.F.-W. Chan, T.T.-T. Yuen, H. Shuai, S. Yuan, Y. Wang, B. Hu, C.C.-Y. Yip, J.O.-L. Tsang, and X. Huang, Lancet Microbe 1(1), e14. (2020).
K.P.Y. Hui, M.-C. Cheung, R.A.P.M. Perera, K.-C. Ng, C.H.T. Bui, J.C.W. Ho, M.M.T. Ng, D.I.T. Kuok, K.C. Shih, and S.-W. Tsao, Lancet Respir. Med. 8(7), 687. (2020).
A.J. Pruijssers, A.S. George, A. Schäfer, S.R. Leist, L.E. Gralinksi, K.H. DinnonIii, B.L. Yount, M.L. Agostini, L.J. Stevens, and J.D. Chappell, Cell Rep. 32(3), 107940. (2020).
J.J. Malin, I. Suárez, V. Priesner, G. Fätkenheuer, and J. Rybniker, Clin. Microbiol. Rev. 34(1), e00162. (2020).
Acknowledgements
NSF Grants #1229316 and #2019077 are acknowledged for the support of the computational resources at NDSU CCAST. The authors also acknowledge the partial support of the National Science Foundation NSF EPSCoR Track-1 Cooperative Agreement OIA #1946202 and the NDSU Grand Challenge funded Center for Engineered Cancer Testbeds.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors indicate that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Faisal, H.M.N., Katti, K.S. & Katti, D.R. Differences in Interactions Within Viral Replication Complexes of SARS-CoV-2 (COVID-19) and SARS-CoV Coronaviruses Control RNA Replication Ability. JOM 73, 1684–1695 (2021). https://doi.org/10.1007/s11837-021-04662-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11837-021-04662-6