Abstract
We investigated the functional and selective activity of three phage-derived gold-binding peptides on the Escherichia coli (E. coli) bacterial cell surface display scaffold (eCPX) for the first time. Gold-binding peptides, p3-Au12 (LKAHLPPSRLPS), p8#9 (VSGSSPDS), and Midas-2 (TGTSVLIATPYV), were compared side-by-side through experiment and simulation. All exhibited strong binding to an evaporated gold film, with approximately a 4-log difference in binding between each peptide and the control sample. The increased affinity for gold was also confirmed by direct visualization of samples using Scanning Electron Microscopy (SEM). Peptide dynamics in solution were performed to analyze innate structure, and all three were found to have a high degree of flexibility. Preferential binding to gold over silicon for all three peptides was demonstrated, with up to four orders of magnitude selectivity exhibited by p3-Au12. The selectivity was also clearly evident through SEM analysis of the boundary between the gold film and silicon substrate. Functional activity of bound E. coli cells was further demonstrated by stimulating filamentation and all three peptides were characterized as prolific relative to control samples. This work shows great promise towards functional and active bacterial–hybrid gold surfaces and the potential to enable the next generation living material interfaces.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The role of biological molecules in the bottom–up assembly of hybrid materials has been recognized as critical in overcoming the challenges of synthetic material construction and the complex, multistep protocols involved therein. In natural systems, molecular recognition and substrate specificity by biological molecules enable basic building blocks to be organized into hierarchical structures that span the atomic scale to macroscale. The integration of biological components (both living organisms and biological molecules), as well as their organization and control, facilitates the hybrid, bottom–up design. This in turn allows for the development of novel system properties and advanced functionality in plasmonics, optics, catalysis, biosensing, power generation and energy storage, among many other related fields.1–5 In particular, the conjugation of biomolecules (proteins, enzymes, deoxyribonucleic acid (DNA), and lipids) as well as living cells to both flat and structured gold has been extensively documented.6–16 Traditionally, biomolecule coupling to gold (or other substrates) is often facilitated by a cross-linking agent and rarely directly to the gold surface itself. Integration of a gold-binding peptide into a biomolecule of interest allows utilization of more complex chemistry and offers the advantage of direct integration with the surface. Furthermore, by exploiting selectivity of integrated peptides for various surfaces, new possibilities are opened for directed assembly and patterning. There is an extensive body of literature devoted to the discovery and study of gold-binding peptides, with minimal investigation of biomolecularly directed bacterial–hybrid interfaces.
Gold-Binding Peptides
Gold-binding peptides are useful molecular linkers due to their robust nature and ease of modification, where functionality is tailored through genetic engineering and synthetic chemistry. Gold-binding peptides are examples of genetically engineered peptides for inorganics (GEPI), and a number of gold-binding peptides have been developed for a variety of applications. A wide range of inorganic materials, including metals, metal oxides, semiconductors, polymers, carbon nanotubes and other functional materials (hydroxylapatite, mica, graphite)17–35 have been the focus of previous studies using GEPI. Traditionally, in such studies, emphasis has been focused on peptide binders with the highest affinity for their target. However, peptides with the strongest target binding are not always desirable. In many applications requiring reversibility, such as biosensors, a moderate binding strength is often preferable. Palafox-Hernandez et al.36 developed a scoring system to estimate peptide binding strength to gold. Using calculations of individual amino acid binding strengths to the aqueous gold interface, a score of two, three, or four (representing weak, medium, or strong, respectively) was assigned for the binding enthalpy contribution. By this criteria, the published gold-binding peptides, p3-Au12, p8#9, and Midas-2 (developed from two types of phage display libraries) were selected for this study and can be considered medium gold binders in strength. These peptides were also chosen because they span a range of physical properties (Table I), as well as primary structural characteristics (e.g., total charge, hydroxyl-richness) while avoiding immobilizing interactions, such as those generated by cysteine.
Peptide p3-Au12 (LKAHLPPSRLPS) was discovered from the commercial type 3 phage display (PhD) library, where the peptides were expressed on the pIII coat protein of filamentous phage, and were screened against the (111) plane of a crystal gold ingot.37 This peptide was later used, in conjunction with a tetraglutamate tag expressed on the pVIII protein, to create gold–cobalt oxide nanowires to improve battery capacity. Peptide p8#9 (VSGSSPDS) was discovered from the type 8 library,38 in which the peptide library was located on the pVIII protein, and was screened for binding to gold thin films after four rounds of sorting.39 Phage particles expressing both the gold-binding peptide and a streptavidin-binding motif were then used to assemble gold and cadmium selenide nanocrystals with complex geometries. This peptide has also been employed to functionalize composite fibers for gold nanoparticle coating,40 to form viral templated gold nanowires for hybrid semiconductor films41 and anode materials in lithium ion batteries.42 Peptide Midas-2 (TGTSVLIATPYV) was developed from a screen against metallic gold powder using the commercial type 3 PhD library.43 The free (i.e., off-cell) Midas-2 peptides were utilized to form monodispersed gold nanoparticles. Although this peptide was not used in other investigations, a derivative peptide, Midas-11, in which the tyrosine at position 11 was replaced with glycine, has been used in a number of follow-up studies.44–46
In addition to the wide variety of gold-binding application studies, there has been a significant effort in the field towards understanding factors and attributing mechanisms behind peptide–gold interactions. Although extensive experimental and computational work has been performed, in part to formulate guidelines in designing gold binders, results have been highly varied and a definitive set of rules has remained elusive. Willett et al. systematically tabulated single amino acid adhesion to a variety of inorganic materials, where it was noted empirically that only amino acid residues Arg, Thr, Asp, Ser, Ile, and Pro displayed much affinity for the non-oxide-forming gold surface.47 Arginine-rich sequences have also been found in the engineered antibody work of Jain et al.48 We note that cysteine is prominently missing from this list, despite the ubiquity of the thiol–gold interaction in the field of self-assembled monolayers (SAMs) and the spectroscopic identification of the sulfur–gold bond in other cysteine-containing gold systems.49 However, relative rankings of amino acids as gold binders in additional work (such as that of Peelle et al.,50 Fears et al.51 and Cohavi52) have not displayed consistent trends.
Additionally, a wealth of computational analysis of gold-binding amino acids and peptides has been performed using a variety of methods. Although much of this work has been designed to isolate properties contributing to successful binding (e.g., flexibility, electrostatics, solvent interactions), it has remained difficult to distill this into a concise set of design principles.36,53,54 In order to probe the origins of the predominance of hydroxyl-containing amino acids with gold surfaces, Calzolari et al.55 performed ab initio molecular dynamics (AIMD) of a hydroxyl containing β-sheet-forming peptide, and found that cooperativity between serine side chains and solvent molecules contributed to successful binding. Hong et al.56 performed a density functional theory (DFT) study of a select group of amino acids interacting with a gold (111) surface and demonstrated the strong effects of charge transfer in binding charged residues such as Asp, Lys, and Arg. Molecular dynamics of the binding process of several gold-binding peptides performed by Yu et al.57 suggested that Tyr, Met, and Phe are strong binders, while serine breaks through the hydration layer to form an anchor point. Verde et al.58 performed further molecular dynamics to study the role of flexibility and stability in adsorption of known gold-binding and non-gold-binding peptides. They found that, in solvated systems, high mobility (configurational sampling and high local flexibility) is necessary to displace the water layer and promote adsorption. This is found to occur in unstructured peptides consisting of random coils, or in peptides with ordered structural elements (such as helices) with flexible connectors. Molecular dynamics performed in conjunction with surface plasmon resonance (SPR) binding experiments by Corni et al.59 on known gold-binding and non-binding peptides corroborated findings by Verde et al.58 and confirmed the importance of peptide flexibility in promoting the binding process. Previous combined experimental/molecular dynamics studies of peptide binding on metal and metal oxide systems demonstrated the important role that solvated peptide conformation plays in the binding process.23,26 Clearly, the large number of factors involved in peptide–surface interactions make this a difficult problem to unravel, both experimentally and computationally.
Surface Display Scaffolds
GEPI are almost exclusively developed from a combinatorial peptide display library, where surface displayed peptides are mined for binding to a specific target. Phage and bacterial cell surface display are the most common libraries used for GEPI biopanning. Phage display typically employs a filamentous bacteriophage with unique peptides (ranging from 5 to 12 amino acids, in both linear and cyclic forms) displayed on one of several coat proteins or on engineered hybrids of natural or artificial coat proteins.60 Several M13 phage display libraries (i.e., p3 library) are commercially available and have been the preferred GEPI library due to their high peptide diversity and the robustness of the viral host to harsh conditions encountered during biopanning with inorganics. Additionally, bacterial surface display libraries have been successfully used to develop GEPI. These are commonly hosted on Escherichia coli (E. coli), with the peptide library located on a modified flagella (FliTrx)61 or modified outer membrane protein (eCPX).23 The eCPX scaffold was the bacterial surface display scaffold used in these studies, and is a circularly permuted form of the OmpX protein that has been optimized to display unconstrained 15mer peptides in E. coli.62,63 The eCPX peptide library has been used previously for biopanning of affinity peptides to several toxins (Protective Antigen component of Anthrax toxin,64,65 Staphylococcal Enterotoxin B (SEB)66), as well as GEPI for a bulk aluminum alloy.23 This scaffold has also been successfully used to display functional peptides developed from phage and allowed for the first direct performance comparison of peptides originating from different scaffolds.23 It was also used to demonstrate the selectivity of an aluminum alloy-binding peptide developed from the eCPX library, for its target alloy over glass, copper or brass.67 Regardless of the peptide display library type or host organisms, the key advantage to all biocombinatorial approaches is that the peptide’s identity is encoded by the organism’s genetic material. Therefore, a direct physical linkage exists between the displayed peptide and the host’s DNA, allowing the amino acid composition to be derived through DNA sequencing analysis. Additionally, the DNA–peptide relationship allows the peptide sequence to be easily altered by re-encoding a portion of the DNA for improved functionality. In many instances, peptides are chemically synthesized after sequence identity discovery for off-cell/off-scaffold applications.
Functional Bacterial–Hybrid Interfaces
It is well known that bacteria naturally attach to surfaces via natural adhesion factors and ultimately form biofilms. Bacterial surface attachment has been exploited and engineered for many biotechnology applications, and a number of cell immobilization techniques have been developed to immobilize cells to an abiotic surface.68,69 However, these techniques are non-specific and rely on general chemical and physical factors including: adsorption, covalent bonding, entrapment, encapsulation and crosslinking.70 For example, Yang et al.71 attached the bacterium Salmonella typhimurium to a gold electrode surface through simple adsorption, while Heiskanen et al.11 conjugated yeast cells to thiol-modified gold microelectrodes. Traditionally, cell attachment to gold, as well as to other surfaces, is not direct, but rather tethered or adhered to the surface through a general physiochemical interaction. Cell immobilization is usually ubiquitous over the surface, or addressed to a particular surface location by masking or through the use of charge effects. On the other hand, peptides inherently have high selectivity in target binding, leading to use of these biomolecules in directed and auto-templated assembly. Peptide specificity is derived through the process of biopanning a combinatorial peptide display library, as iterative rounds of biopanning with an increasing stringency and counter selection steps select for peptides with high affinity and specificity. After the initial on-cell peptide discovery is complete, the peptides are often synthesized off-cell for further characterization and implementation into directed assembly biohybrid systems, and a number of review articles have been devoted to the topic of off-cell peptide-surface interactions.72–74
In contrast, there are very few examples of peptide-directed bacterial interfaces in the literature, with limited investigation of functional integration and viability. Baneyx, Sarikaya, and coworkers have demonstrated compatibility of biocombinatorial discovery and viability in the context of GEPI using the FliTrx E. coli cell surface display scaffold. In their work, they showed selectivity towards monovalent oxide over higher valency films, and demonstrate viability through recovery of E. coli after 15 min of incubation with the cuprous oxide surface.26,75 Most recently, we have shown up to 24 h of viability from a peptide-directed interface between E. coli and an aluminum alloy employing the eCPX surface display scaffold.23 Herein, we demonstrate the ability to express functional, phage-derived gold-binding peptides, in a cross-scaffold manner on the E. coli bacterial cell surface display scaffold, eCPX. Through this approach, we include a side-by-side comparison of peptides and their facilitated cell binding to gold for the first time. Investigations include demonstration of gold-binding affinity, and peptide selectivity for gold over silicon, as well as a discussion of these interactions incorporating flexibility through molecular dynamics and strength of interactions through experimental study. Finally, we demonstrate functional and active bacterial–hybrid gold surfaces for the first time, showing great promise for next generation living material interfaces.
Materials and Methods
Bacterial Strains, Culture Conditions, and Materials
E. coli MC1061 (Lucigen, Middleton, WI, USA) was routinely maintained in LB Miller broth (Becton, Dickinson and Company, Franklin Lakes, NJ USA) supplemented with 25 µg/mL chloramphenicol at 37°C, 225 rpm. Phage-derived gold-binding peptides, p3-Au12,37 p8#9,39 and Midas-243 were synthesized (BioBasic, Amherst, NY, USA) for cloning into the eCPX vector as previously described.23 The resulting plasmids were transformed into chemically competent MC1061 cells and the peptide sequences verified by sequencing (Genewiz, South Plainfield, NJ, USA).
Gold was evaporated onto a standard 4 inch (c. 10 cm) silicon wafer at a surface thickness of 200 nm deposited at 2–3 Å/s with a 50 nm titanium or chromium adhesion layer deposited at 2–3 Å/s and cleaned with hydrofluoric acid. The surface was analyzed by x-ray powder diffraction (XRD) and found to be predominately a (111) surface. The gold and silicon wafers were then cleaved into strips sized approximately 10 mm × 30 mm. The wafer samples were sterilized prior to use by exposure to a germicidal UV lamp for 30 min on each side.
All molecular and microbiology support materials (e.g., primers, buffers, enzymes, media, Tween20, antibiotics, etc.) were obtained from standard, commercial suppliers (Fisher Scientific, Sigma Aldrich, Invitrogen, NEB, etc.) and used according to standard techniques.
Indirect Binding Assay
Indirect binding assays were used to quantify the number of cells bound to both the gold and silicon wafers and were performed as previously described.23 Briefly, the material was incubated with an E. coli culture displaying either a gold-binding peptide or the eCPX scaffold only. After washing to remove weakly bound cells, the material was transferred to LB supplemented with 0.2% glucose to recover those bound cells, and serial dilutions were plated to determine the number of cells bound in each material set.
Scanning Electron Microscopy
Scanning Electron Microscopy (SEM) imaging was performed under high vacuum using a FEI Quanta 200F environmental SEM (accelerating voltage = 2 keV). For direct quantification of cells on the material surface, cell incubation and washing was performed as previously described,23 except that after washing the samples were allowed to fully dry (typically overnight under ambient laboratory conditions) before imaging. Samples were attached to aluminum stubs using carbon tape. For the cell density estimates, ten regions (area of 750 μm2 each) were imaged at random on the substrate. Cells were enumerated to determine the overall cell density, or cell surface coverage was estimated in cases where the cell density was too high for direct cell counts. In that case, the brightness contrast between the substrate and cells was analyzed using the image processing capabilities of the XT microscope control software.
Cell Activity on Gold
The viability of cells bound to gold through each respective gold-binding peptide was achieved by inhibiting cell cleavage after division by the addition of the antibiotic, aztreonam. Thus, viable cells filamented into chains and dead cells remained as single cells. After binding and washing, as described above, gold wafers were placed in LB supplemented with 25 µg/mL chloramphenicol, 0.04% arabinose, and 20 µg/mL aztreonam, a sub-lethal concentration previously determined cause filamentation in this strain, for 7 h at 37°C with shaking. A second set of samples were incubated in LB supplemented with only 25 µg/mL chloramphenicol and 0.04% arabinose to serve as a control. After the 7 h incubation, the gold samples were removed and dried overnight for imaging by SEM.
Statistical Analysis
Either an unpaired t tests or one-way ANOVA were preformed where appropriate using Prism Graph Pad 5 software with the included statistical package. Statistical significance was defined as p < 0.05.
Molecular Dynamics
Molecular dynamics simulations were performed on each peptide (p3-Au12, p8#9, and Midas-2) using the NAMD software of Schulten and co-workers.76 VMD was used for simulation system setup by a three-step process involving peptide construction via the Molefacture plugin, solvation with a 15 Å TIP3 water buffer on each edge of the peptide structure, and ionization with NaCl to achieve overall simulation box neutrality. As an indication of total system size, for the p3-Au12 peptide this resulted in simulation cell of 62.46 Å × 46.88 Å × 41.94 Å and a total of 11,317 atoms. The system was sequentially minimized and heated to a final temperature of 300 K over 4500 steps. NPT dynamics were performed using the CHARMM forcefield with hydrogen bonds held rigid using the SHAKE algorithm, a timestep of 2 fs, and pressure of 1 atm. The simulation was performed for 40 ns, representing 10 ns of equilibration and 30 ns of production. RMSD analysis was performed with the RMSD Trajectory Tool (RMSDTT) plugin in VMD.
Results and Discussion
Comparison of Peptide Binding to Gold
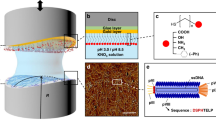
The three peptides featured in this work were discovered using two types of phage display libraries. The peptide p8#9 was developed from a type 8 library, whereas p3-Au12 and Midas-2 were developed from a type 3 library. Although these peptides were discovered and originally characterized on phage, it has been documented that cross-phage scaffold peptide function is poor for phage coat proteins.39,60,77 Therefore, the peptides were displayed on the eCPX display scaffold, allowing for a side-by-side comparison of functional activity on the E. coli surface. This cell surface display scaffold has been previously used to successfully display functional phage-derived peptides.23 After cloning each peptide into the N-terminus of eCPX, binding to a gold wafer was measured using an indirect cell-binding assay shown in Fig. 1. All three peptides were found to strongly bind to the gold wafer, compared to the negative control, exhibiting almost a 4-log difference in binding between each peptide and the negative control (eCPX scaffold alone expressed to the same level). The number of cells recovered from the gold wafer was adjusted for wafer sample size, and cell binding for all three peptides were on the order of 106 cells/mL cm2. By contrast, the negative control showed cell binding on the order of 101 cells/mL cm2 (Fig. 1) under the same conditions. These results confirm that phage derived peptides were successfully displayed and functional on the E. coli eCPX scaffold. Furthermore, the large difference in gold binding between cells expressing a gold-binding peptide and the negative control (no peptide scaffold) demonstrates that gold binding is specifically facilitated by the presence of the peptides and not due to non-specific interactions between cell surface components and the gold wafer. Overall, a comparison of the density of gold-bound cells for each of the peptides shown in Fig. 1 indicates that there was a statistically significant (p < 0.05) difference between peptide p8#9 and Midas-2. There was no significant difference in gold binding between p8#9 and p3-Au12, and p3-Au12 and Midas-2. These results suggest that the p8#9 may have a higher affinity for gold on the eCPX scaffold, although all three peptides bind well to gold.
It was not surprising that p8#9 facilitated cell binding to the gold surface at the highest level among the three peptides examined. This peptide was discovered from biopanning using a thin gold film,39 and it is likely that this material was very similar to the gold wafer used in the studies reported herein. It is likely that both materials were polished, gold surfaces. The target used to discover peptide p3-Au12 was a (111) plane of a crystal gold ingot37 and cells displaying this peptide were found to bind the gold wafer only slightly less than p8#9. It is possible that p3-Au12 may be more specific for gold (111), which may have impacted the gold binding to the wafer. Midas-2, the peptide displayed on cells that bound the least well to the gold wafer, as it was discovered against a gold powder.43 Gold powder, although chemically identical, is structurally very different than a gold film and therefore it is difficult to anticipate relative binding interactions. However, it is interesting to note that Midas-2 also exhibited binding to a gold thin film.
The increased affinity for gold with the expression of gold-binding peptides on the cell surface was also confirmed by direct visualization of samples using SEM. Figure 2 shows representative images of gold surfaces before use (Fig. 2a), after incubation with the negative control (Fig. 2b), and cells displaying the p8#9 gold-binding peptide (Fig. 2c). SEM images of gold surfaces bound by cells displaying the two other gold-binding peptides appeared similar to Fig. 2c. Briefly, images were taken across the gold surface and cell counts from these images indicated that very few cells were present in samples incubated with the negative control (<103 cells/cm2), while at least 105 cells/cm2 were present on samples incubated with cells expressing the gold-binding peptide. It should be noted that, because cell binding was enumerated directly on the wafer by SEM, these samples were not recovered in culture media after washing and instead were immediately imaged. Even with that distinction in sample processing, the direct cell counts by microscopy were comparable to cell counts obtained from the indirect binding assay.
Peptide dynamics in solution were studied to analyze innate structure and flexibility, which have been shown to play an important role in the binding process.26,57,58,67,78 It is clear from Fig. 3 that all three peptides are lacking in helical propensity and avoid any overwhelming structural characteristics which could interfere in the binding process. The most noteworthy structural characteristic is the two-proline bend in the middle of p3-Au12 (Fig. 3a). However, the segments on either side of this bend are completely flexible and this small structure is not expected to impede the binding process in any meaningful way. It is possible that these proline residues contribute to the ‘dynamic anchoring’ seen by Yu et al.57 An analysis of the backbone RMSD during the course of a 30 ns trajectory allows comparison of the overall RMSD for each peptide, showing that while Midas-2 is somewhat more flexible, all three peptides show a similar degree of flexibility. These similarities in flexibility and lack of innate structure are consistent with our experimental results, which do not discriminate binding on gold between these three systems. The Midas-2 structures are also similar to previously published data of this peptide binding on gold,57 which highlight the flexible middle Val-Leu-Ile-Ala segment (clearly visible in Fig. 3c) and the possible role it plays in the binding process. Updated dynamics simulations of each of these systems on the gold (111) surface is currently underway.
Selectivity of Gold-Binding Peptides
To fully utilize the potential of GEPI, it is important to not only gain insight into its affinity to its target inorganic but to also characterize its specificity such that it may be used successfully in other applications and tailored for other functionalities. Knowledge of a peptide’s specificity to a given material compared to other materials is critical for the use of peptides in complex, mixed material systems.79 In fact, it can be argued that GEPI selectivity is more important than overall target affinity for incorporation into hybrid structures and devices. In order to gain some insight into the selectivity of the three phage-derived gold-binding peptides, indirect binding assays were conducted to determine cell binding to gold and silicon substrates. There was a statistically significant (p < 0.05) preference of the three peptides for gold over silicon, as shown in Fig. 4. Peptide p3-Au12 had the greatest difference in binding to gold over silicon at 4 orders of magnitude. Peptides p8#9 and Midas-2 were found to have a lesser degree of selectivity for gold over silicon, at 1 order of magnitude, but were still considered to be selective for gold. Gold-binding peptide selectivity was directly visualized by examining the boundary between the evaporated gold layer and the uncoated silicon wafer by SEM. Representative images are shown in Fig. 5, and clearly show the preference of cells displaying gold-binding peptide p3-Au12 for the gold-coated surface. Similar images were obtained for cells displaying the other gold-binding peptides. The gold-coated region of the wafer contained a high density of bound cells; however, at the interface of the gold and silicon, the density of bound cells decreased. Approximately 100 µm beyond the transition from gold to silicon, there was little or no cell binding to the silicon. Even after 7 h in culture media, cells displaying a gold-binding peptide showed a very strong preference for binding to gold over silicon. An estimation of cell density on the two regions found that peptide-mediated cell binding was approximately 45–60% coverage on gold and approximately 0.1% coverage on silicon. This demonstrates that these peptides can be used to selectively seed a biofilm, where there is significant cell attachment to the target material, gold in this case, and virtually no attachment to the underlying background substrate.
As binding was tested against sections of gold-coated silicon wafer, the dull silicon back surface of the wafer was therefore also exposed and may potentially contribute to recovered cell counts from the indirect binding assay. Therefore, both the polished front side and the dull back side of uncoated silicon wafers were analyzed for cell binding by SEM. There was essentially no cell binding to the polished front side; however, there was observable displayed peptide cell binding to the dull, back side surface. Cell counts were approximately 1 order of magnitude less than binding to the gold. These data indicate that non-specific background binding does occur to the unpolished silicon surface; however, it does not significantly contribute the cell counts obtained from the indirect binding assay with the displayed peptides on gold.
Peptide selectivity to inorganic materials has been the focus of several investigations and reviews, although the mechanisms by which a peptide selectively binds to one metal surface over the oxide or between two similar metals is not well understood.79 Peptide selectivity is likely due to the recognition of a combination of chemical (hydrogen bonding, polarity, and charge effects) and structural (size and morphology) features.80,81 Whaley et al.29 described a peptide that was specific not only to gallium arsenide over silicon, but bound the (100) crystal face over the (111)B. The specificity of another gold binding peptide, GBP1, was examine using a quartz-crystal microbalance and was found to preferentially absorb to gold over platinum and silica, and suggested that polar moieties and the physical conformation of the peptide itself may play a role in the binding preference to gold over platinum or silica.79
While clear trends from the literature remain elusive, a simple analysis of the three-peptide sequences in the context of known gold and silicon binders was performed. If one examines the sequences for gold-binding residues based on the work of Willett and colleagues47 (Arg, Thr, Asp, Ser, Ile, Pro), then all peptides are roughly equivalent at six potential gold-binding residues each. As determined from molecular dynamics (Fig. 3), the flexibility of each of the peptides is roughly equivalent, so this is also not a discriminating factor, nor is there any inherent structure to these systems to influence how binding moieties present to the oncoming surface. It is possible that the slightly improved affinity of peptide p8#9 for gold may be attributed to the larger number of serines in this peptide. It has been previously noted55,57 that the interactions of hydroxyl-containing residues play an important role in providing access to the surface, thus allowing opportunity for binding. Less is known about silicon-specific binding residues in the literature. The doping of silicon systems, which are vital to their properties as semiconductors, further complicates this analysis. However, Estephan et al.27 developed peptide binders for n+, p and p+ silicon surfaces, and a subset of these has been studied by molecular dynamics.82 It is noteworthy that the binders developed for p and p+ silicon contain a high number of hydroxyl (Ser, Thr, Tyr) and carboxyl acid (Asp, Glu) groups in their sequences. This is not surprising in the context of small molecule adsorption on silicon surfaces, where oxygen-containing functional groups (such as hydroxyl groups and carboxylic acids) have been recognized for their role in forming silicon-oxygen bonds.83 Again, a very simple analysis of the sequences of our three peptides, based upon these observations, shows that p8#9 has four hydroxyl-containing residues and an acidic residue, while Midas-2 has five hydroxyl-containing residues. This can be attributed to the lesser selectivity for gold displayed by these peptides. By contrast, p3-Au12, has only two hydroxyl-containing residues, does not bind silicon as well, and demonstrates a higher degree of selectivity.
Activity of Cells Bound to Gold
The incorporation of free peptides into inorganic systems requires peptides to have high affinity and good specificity for the target material. Due to the inherent robust nature of peptides, the effect of the surface on the integrity of the peptide has not been a significant concern. However, biohybrid material systems will likely incorporate living functionalized cells to perform a biological process while specifically bound to a given material. In order to have cellular performance in biohybrid systems, bound cells must remain viable and metabolically active while on their target substrate. For this reason, we showed that the E. coli cells bound to the gold wafer via each of three gold-binding peptides remained active, as demonstrated by their ability to divide. The addition of a sublethal concentration of the antibiotic, aztreonam, allowed the cells to divide but prevented the cell wall from separating, leading to filamentation.84 Therefore, non-dividing cells had a typical cell length (~2 µm) and active, dividing cells filamented into chains 50 µm or longer. After incubating cells displaying each of the three peptides with the gold wafer and washing, the samples were placed in culture media supplemented with aztreonam and arabinose, to facilitate the binding of newly filamented cells to the gold surface. After samples were removed and dried, cells were imaged by SEM, as shown in Fig. 6. There was notable filamentation in all samples incubated with aztreonam, while no filamentation was observed in control samples (no aztreonam) (data not shown). Extensive filamentation was seen in cells displaying all three peptides on the gold surface. Although the high cell density prevented the measurement of accurate filament length, we can estimate that a significant portion of the filaments approach 100 µm or longer. Cells displaying peptides p8#9 (Fig. 6b) and p3-Au12 (Fig. 6d) both had the greatest amount of surface coverage and formed the longest filaments. This is consistent with these peptides having the best binding to gold. Cells displaying peptide Midas-2 (Fig. 6c) had a lower level of surface coverage and had shorter filaments. This was still greater than the sample containing the negative control cells (Fig. 6a), which were sparse and had the lowest degree of filamentation. For this sample, the cell density was calculated to be approximately 102 to 103 cells/cm2, values consistent with the binding data presented in Fig. 1. It should be noted that cell densities could not be accurately determined for samples with the three gold-binding peptides due to overlapping filaments, but can be estimated to be approximately three orders of magnitude higher than the negative control. Single cells were observed in all samples, indicating that dead or dying cells were present. This is likely due to the length of time cells were in the culture medium (7 h) and are not believed to be due to adverse effects of binding to the gold surface. Taken together, these result clearly demonstrate that cells bound to the gold surface remained active, and binding to gold did not appear to compromise cell viability, activity, or replication in a significant manner.
Conclusion
Gold and silicon are two of the most common inorganic materials utilized in electronic processing applications. Therefore, when considering generation and control of biohybrid functional surfaces, material selectivity is paramount as we move beyond simple, non-specific methodologies (e.g., thiol chemistry or hydrogels, etc.) In this work, we showed, for the first time, successful cross-scaffold integration of three gold-binding peptides, with widely varying physical properties, yet notably lacking in cysteine and tyrosine residues. All three peptide-directed cell constructs were found to strongly and selectively bind to gold over silicon. This highlights the need to further the understanding of peptide–target interactions by looking beyond obvious surface chemistries and considering other contributing factors such as flexibility, structure and solvation effects. An important consideration towards biohybrid functional surfaces integrating living organisms is viability and activity. In this work, we demonstrated, for the first time, auto-cell templating directed by a selective peptide interface, and a resulting highly active, biohybrid material, through controlled cell filamentation. This work shows great promise towards functional and active bacterial–hybrid gold surfaces, and future work will seek to further understand and explore the potential of next generation living material interfaces for game-changing technologies.
References
L. Xu, H. Kuang, C. Xu, W. Ma, L. Wang, and N.A. Kotov, J. Am. Chem. Soc. 134, 1699 (2012).
K.T. Nam, D.-W. Kim, P.J. Yoo, C.-Y. Chiang, N. Meethong, P.T. Hammond, Y.-M. Chiang, and A.M. Belcher, Science 312, 885 (2006).
D. Aili, P. Gryko, B. Sepulveda, J.A. Dick, N. Kirby, R. Heenan, L. Baltzer, B. Liedberg, M.P. Ryan, and M.M. Stevens, Nano Lett. 11, 5564 (2011).
K. Zhu, D. Wang, and J. Liu, Nano Res. 2, 1 (2009).
R.M. Kong, X.B. Zhang, Z. Chen, and W. Tan, Small 7, 2428 (2011).
A.J. Di Pasqua, R.E. Mishler, Y.-L. Ship, J.C. Dabrowiak, and T. Asefa, Mater. Lett. 63, 1876 (2009).
H. Cho, J. Zook, T. Banner, S.-H. Park, B.-H. Min, K.A. Hasty, E. Pinkhassik, and E. Lindner, Tissue Eng. Regen. Med. 11, 10 (2014).
D. Athey, M. Ball, C.J. Mcneil, and R.D. Armstrong, Electroanalysis 7, 270 (1995).
S. Zhang, N. Wang, H. Yu, Y. Niu, and C. Sun, Bioelectrochemistry 67, 15 (2005).
O. Shulga and J.R. Kirchhoff, Electrochem. Commun. 9, 935 (2007).
A.R. Heiskanen, C.F. Spegel, N. Kostesha, T. Ruzgas, and J. Emnéus, Langmuir 24, 9066 (2008).
L. Tong, Q. Wei, A. Wei, and J.X. Cheng, Photochem. Photobiol. 85, 21 (2009).
L.M. Demers, M. Östblom, H. Zhang, N.-H. Jang, B. Liedberg, and C.A. Mirkin, J. Am. Chem. Soc. 124, 11248 (2002).
A. Csáki, P. Kaplanek, R. Möller, and W. Fritzsche, Nanotechnology 14, 1262 (2003).
B. Cornell, V. Braach-Maksvytis, L. King, P. Osman, B. Raguse, L. Wieczorek, and R. Pace, Nature 387, 580 (1997).
C.S. Thaxton, W.L. Daniel, D.A. Giljohann, A.D. Thomas, and C.A. Mirkin, J. Am. Chem. Soc. 131, 1384 (2009).
M. Sarikaya, C. Tamerler, A.K.-Y. Jen, K. Schulten, and F. Baneyx, Nat. Mater. 2, 577 (2003).
S. Brown, Nat. Biotechnol. 15, 269 (1997).
S. Brown, M. Sarikaya, and E. Johnson, J. Mol. Biol. 299, 725 (2000).
R.R. Naik, L.L. Brott, S.J. Clarson, and M.O. Stone, J. Nanosci. Nanotechnol. 2, 95 (2002).
K. Kjærgaard, J.K. Sørensen, M.A. Schembri, and P. Klemm, Appl. Environ. Microbiol. 66, 10 (2000).
R. Zuo, D. Örnek, and T.K. Wood, Appl. Microbiol. Biotechnol. 68, 505 (2005).
B.L. Adams, A.S. Finch, M.M. Hurley, D.A. Sarkes, and D.N. Stratis-Cullum, Adv. Mater. 25, 4585 (2013).
R.R. Naik, S.J. Stringer, G. Agarwal, S.E. Jones, and M.O. Stone, Nat. Mater. 1, 169 (2002).
E. Estephan, C. Larroque, F.J. Cuisinier, Z. Bálint, and C. Gergely, J. Phys. Chem. B 112, 8799 (2008).
C.K. Thai, H. Dai, M. Sastry, M. Sarikaya, D.T. Schwartz, and F. Baneyx, Biotechnol. Bioeng. 87, 129 (2004).
E. Estephan, M.B. Saab, M. Martin, C. Larroque, F. Cuisinier, O. Briot, S. Ruffenach, M. Moret, and C. Gergely, J. Pept. Sci. 17, 143 (2011).
C.E. Flynn, C. Mao, A. Hayhurst, J.L. Williams, G. Georgiou, B. Iverson, and A.M. Belcher, J. Mater. Chem. 13, 2414 (2003).
S.R. Whaley, D. English, E.L. Hu, P.F. Barbara, and A.M. Belcher, Nature 405, 665 (2000).
A.B. Sanghvi, K.P.-H. Miller, A.M. Belcher, and C.E. Schmidt, Nat. Mater. 4, 496 (2005).
S. Wang, E.S. Humphreys, S.-Y. Chung, D.F. Delduco, S.R. Lustig, H. Wang, K.N. Parker, N.W. Rizzo, S. Subramoney, and Y.-M. Chiang, Nat. Mater. 2, 196 (2003).
M.D. Roy, S.K. Stanley, E.J. Amis, and M.L. Becker, Adv. Mater. 20, 1830 (2008).
C. Li, G.D. Botsaris, and D.L. Kaplan, Cryst. Growth Des. 2, 387 (2002).
C.R. So, Y. Hayamizu, H. Yazici, C. Gresswell, D. Khatayevich, C. Tamerler, and M. Sarikaya, ACS Nano 6, 1648 (2012).
E.M. Krauland, B.R. Peelle, K.D. Wittrup, and A.M. Belcher, Biotechnol. Bioeng. 97, 1009 (2007).
J.P. Palafox-Hernandez, Z. Tang, Z.E. Hughes, Y. Li, M.T. Swihart, P.N. Prasad, T.R. Walsh, and M.R. Knecht, Chem. Mater. 26, 4960 (2014).
D.J. Solis, Academic Massachusetts Institute of Technology Thesis (2006).
S.-K. Lee, D.S. Yun, and A.M. Belcher, Biomacromolecules 7, 14 (2006).
Y. Huang, C.-Y. Chiang, S.K. Lee, Y. Gao, E.L. Hu, J.D. Yoreo, and A.M. Belcher, Nano Lett. 5, 1429 (2005).
C.Y. Chiang, C.M. Mello, J. Gu, E.C. Silva, K.J. Van Vliet, and A.M. Belcher, Adv. Mater. 19, 826 (2007).
E.D. Haberer, J.H. Joo, J.F. Hodelin, and E.L. Hu, Nanotechnology 20, 415206 (2009).
Y.J. Lee, Y. Lee, D. Oh, T. Chen, G. Ceder, and A.M. Belcher, Nano Lett. 10, 2433 (2010).
J. Kim, Y. Rheem, B. Yoo, Y. Chong, K.N. Bozhilov, D. Kim, M.J. Sadowsky, H.-G. Hur, and N.V. Myung, Acta Biomater. 6, 2681 (2010).
S.-H. Shin, G.-Y. Kim, J. Shim, J. Kim, H.-G. Hur, D.-J. Lee, J.-I. Song, and S.-H. Moon, Korean J. Chem. Eng. 29, 1666 (2012).
J.M. Galloway and S.S. Staniland, J. Mater. Chem. 22, 12423 (2012).
K.-Y. Tomizaki, S. Wakizaka, Y. Yamaguchi, A. Kobayashi, and T. Imai, Langmuir 30, 846 (2014).
R. Willett, K. Baldwin, K. West, and L. Pfeiffer, Proc. Natl. Acad. Sci. USA. 102, 7817 (2005).
P. Jain, A. Soshee, S.S. Narayanan, J. Sharma, C. Girard, E. Dujardin, and C. Nizak, J. Phys. Chem. A 118, 14502 (2014).
S. Aryal, B. Remant, N. Dharmaraj, N. Bhattarai, C.H. Kim, and H.Y. Kim, Spectrochim. Acta A 63, 160 (2006).
B.R. Peelle, E.M. Krauland, K.D. Wittrup, and A.M. Belcher, Acta Biomater. 1, 145 (2005).
K.P. Fears, T.D. Clark, and D.Y. Petrovykh, J. Am. Chem. Soc. 135, 15040 (2013).
O. Cohavi, D. Reichmann, R. Abramovich, A.B. Tesler, G. Bellapadrona, D.B. Kokh, R.C. Wade, A. Vaskevich, I. Rubinstein, and G. Schreiber, Chem. Eur. J. 17, 1327 (2011).
J. Feng, R.B. Pandey, R.J. Berry, B.L. Farmer, R.R. Naik, and H. Heinz, Soft Matter 7, 2113 (2011).
M. Hoefling, F. Iori, S. Corni, and K.-E. Gottschalk, Langmuir 26, 8347 (2010).
A. Calzolari, G. Cicero, C. Cavazzoni, R. Di Felice, A. Catellani, and S. Corni, J. Am. Chem. Soc. 132, 4790 (2010).
G. Hong, H. Heinz, R.R. Naik, B.L. Farmer, R. Pachter, and A.C.S. Appl, Mater. Interfaces 1, 388 (2009).
J. Yu, M.L. Becker, and G.A. Carri, Langmuir 28, 1408 (2011).
A.V. Verde, J.M. Acres, and J.K. Maranas, Biomacromolecules 10, 2118 (2009).
S. Corni, M. Hnilova, C. Tamerler, and M. Sarikaya, J. Phys. Chem. C 117, 16990 (2013).
V.A. Petrenko and G.P. Smith, Vectors and modes of display, ed S.S. Sidhu and C.R. Geyer (Boca Raton, FL: CR Press, 2005), pp. 63–110.
F. Baneyx and D.T. Schwartz, Curr. Opin. Biotechnol. 18, 312 (2007).
J.J. Rice and P.S. Daugherty, Protein Eng. Des. Sel. 21, 435 (2008).
J.J. Rice, A. Schohn, P.H. Bessette, K.T. Boulware, and P.S. Daugherty, Protein Sci. 15, 825 (2006).
J.M. Kogot, Y. Zhang, S.J. Moore, P. Pagano, D.N. Stratis-Cullum, D. Chang-Yen, M. Turewicz, P.M. Pellegrino, A. De Fusco, H.T. Soh, and N.E. Stagliano, PLoS One 6, e26925 (2011).
D.A. Sarkes, B.L. Dorsey, and D.N. Stratis-Cullum, SPIE Defense, Security, and Sensing (2015).
J.M. Pennington, J.M. Kogot, D.A. Sarkes, P.M. Pellegrino, and D.N. Stratis-Cullum, SPIE Defense, Security, and Sensing (2012).
B.L. Adams and D.N. Stratis-Cullum, Emerg. Mater. Res. 4, 297 (2015).
M.M. Elnashar, J. Biomater. Nanobiotechnol. 1, 61 (2010).
V. Nedovic and R. Willaert, Fundamentals of Cell Immobilisation Biotechnology, Series (Dordrecht: Springer, 2013), p. 493.
G.F. Bickerstaff Jr, Immobilization of Enzymes and Cells, ed J.M. Walker (Totowa, NJ:Humana Press, 1997), pp. 1–11.
L. Yang, C. Ruan, and Y. Li, Biosens. Bioelectron. 19, 495 (2003).
A. Vallee, V. Humblot, and C.-M. Pradier, Acc. Chem. Res. 43, 1297 (2010).
U.O.S. Seker and H.V. Demir, Molecules 16, 1426 (2011).
B.T. Karaca, M. Hnilova, and C. Tamerler, Addressable Biological Functionalization of Inorganics: Materials-Selective Fusion Proteins in Bio-nanotechnology, ed M.R. Knecht and T.R. Walsh (New York: Springer, 2014), pp. 221–255.
H. Dai, C.K. Thai, M. Sarikaya, F. Baneyx, and D.T. Schwartz, Langmuir 20, 3483 (2004).
J.C. Phillips, R. Braun, W. Wang, J. Gumbart, E. Tajkhorshid, E. Villa, C. Chipot, R.D. Skeel, L. Kale, and K. Schulten, J. Comput. Chem. 26, 1781 (2005).
Y.J. Lee and K.T. Nam, Engineered Biomolecules as Nanomaterials, ed D.K. Yi and G.C. Papaefthymiou (Boco Raton, FL: CRC Press, 2013), pp. 287–340.
F. Causa, R. Della Moglie, E. Iaccino, S. Mimmi, D. Marasco, P. Scognamiglio, E. Battista, C. Palmieri, C. Cosenza, and L. Sanguigno, J. Colloid Interface Sci. 389, 220 (2013).
C. Tamerler, M. Duman, E.E. Oren, M. Gungormus, X. Xiong, T. Kacar, B.A. Parviz, and M. Sarikaya, Small 2, 1372 (2006).
S. Mann, B.R. Heywood, S. Rajam, and J.D. Birchall, Nature 334, 692 (1988).
I. Weissbuch, L. Addadi, and L. Leiserowitz, Science 253, 637 (1991).
S.K. Ramakrishnan, M. Martin, T. Cloitre, L. Firlej, and C. Gergely, J. Chem. Inf. Comput. Sci. 54, 2117 (2014).
M.P. Casaletto, R. Zanoni, M. Carbone, M.N. Piancastelli, L. Aballe, K. Weiß, and K. Horn, Surf. Sci. 505, 251 (2002).
S.R. Arends and D.S. Weiss, J. Bacteriol. 186, 880 (2004).
Acknowledgements
The authors thank Dr. Justin Bickford at US Army Research Laboratory for preparation of the gold film substrates utilized in this work, and Drs. Hong Dong and Dat Tran at US Army Research Laboratory for characterization of the prepared gold films by XRD. The authors also thank Ms. Deborah Sarkes at US Army Research Laboratory for laboratory technical support and assistance with technical editing of this manuscript. This project was supported in part by appointments to the US Army Research Laboratory Postdoctoral Fellowship Program administered by the Oak Ridge Associated Universities through a contract with the US Army Research Laboratory.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Adams, B.L., Hurley, M.M., Jahnke, J.P. et al. Functional and Selective Bacterial Interfaces Using Cross-Scaffold Gold Binding Peptides. JOM 67, 2483–2493 (2015). https://doi.org/10.1007/s11837-015-1662-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11837-015-1662-7