Abstract
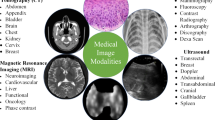
Medical image analysis helps in resolving clinical issues by examining clinically generated images. In today’s world of deep learning (DL) along with advances in computer vision, the available medical imaging data from scientific archives, academic publications, and clinical manuals along with advances in computer vision, present an opportunity could be of great use. The implication of artificial intelligence and DL has increased steadily over the past decades. Studies based on convolutional neural networks (CNNs) are the primary focus, as it is often regarded as one of the most effective tools for analyzing images and other structured data. CNN models naturally understand space and hierarchy using backpropagation, convolution, pooling, and link layers. Most of the research is concentrated on image pre-processing, classification, and evaluating various applications of CNN techniques in medical imaging. To explore this field of research, we carried out a survey wherein we analyzed the papers that were pertinent to MRI image analysis, specifically MRI pre-processing, segmentation and diagnosis. This article provides a thorough overview of medical image analysis by CNNs, along with its shortcomings and potential. The application of CNNs in medical image analysis, such as its use for brain magnetic resonance imaging (MRI) analysis, pre-processing, segmentation, data preparation, and post-processing were the main areas of focus. We further explored large-scale retrieval, which can increase MRI image processing efficiency. Overall, this study analyzed current advancements in this area and the associated major challenges. The publications were categorized by pattern recognition applications before classifying them using a taxonomy based on human architecture.






Similar content being viewed by others
References
Liu X et al (2019) A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: a systematic review and meta-analysis. Lancet Digit Health 1(6):e271–e297. https://doi.org/10.1016/S2589-7500(19)30123-2
Ma J, Song Y, Tian X, Hua Y, Zhang R, Wu J (2020) Survey on deep learning for pulmonary medical imaging. Front Med 14(4):450–469. https://doi.org/10.1007/s11684-019-0726-4
Huang L, Peng J, Zhang R, Li G, Lin L (2018) Learning deep representations for semantic image parsing: a comprehensive overview. Front Comput Sci 12(5):840–857. https://doi.org/10.1007/s11704-018-7195-8
Zhao C, Sun Q, Zhang C, Tang Y, Qian F (2020) Monocular depth estimation based on deep learning: an overview. Sci China Technol Sci 63(9):1612–1627. https://doi.org/10.1007/s11431-020-1582-8
Zhou A, Cui Y, Jiang T (2018) Multisite Schizophrenia classification based on brainnetome atlas by deep learning. In: 2018 5th IEEE international conference on cloud computing and intelligence systems (CCIS), pp 451–455. https://doi.org/10.1109/CCIS.2018.8691336.
Quintana Y, Safran C (2017) Chapter 1—global health informatics—an overview. In: de Fátima Marin H, Massad E, Gutierrez MA, Rodrigues RJ, Sigulem D (eds) Global Health Informatics. Academic Press, New York, pp 1–13. https://doi.org/10.1016/B978-0-12-804591-6.00001-X
Asgari Taghanaki S, Abhishek K, Cohen JP, Cohen-Adad J, Hamarneh G (2021) Deep semantic segmentation of natural and medical images: a review, vol 54, no 1. Springer, Netherlands. https://doi.org/10.1007/s10462-020-09854-1.
Sinha GR (2018) Research studies on human cognitive ability K. Srujan Raju and Raj Kumar Patra Daw Win Aye and Daw Thuzar Khin’ 5(4):298–304
Subudhi A, Sahoo S, Biswal P, Sabut S (2018) Segmentation and classification of ischemic stroke using optimized features in brain MRI. Biomed Eng 30(03):1850011. https://doi.org/10.4015/S1016237218500114
Greenspan H, van Ginneken B, Summers RM (2016) Guest editorial deep learning in medical imaging: overview and future promise of an exciting new technique. IEEE Trans Med Imaging 35(5):1153–1159. https://doi.org/10.1109/TMI.2016.2553401
Lundervold AS, Lundervold A (2019) An overview of deep learning in medical imaging focusing on MRI. Z Med Phys 29(2):102–127. https://doi.org/10.1016/j.zemedi.2018.11.002
Sutskever I, Vinyals O, Le QV (2014) Sequence to sequence learning with neural networks. Adv Neural Inf Process Syst 4:3104–3112
Cheng A et al (2019) Deep learning image reconstruction method for limited-angle ultrasound tomography in prostate cancer. In: Medical imaging 2019: ultrasonic imaging and tomography, vol 10955, pp 256–263. https://doi.org/10.1117/12.2512533
Li Y et al (2021) A comprehensive review of markov random field and conditional random field approaches in pathology image analysis, no. 0123456789. Springer, Netherlands. https://doi.org/10.1007/s11831-021-09591-w
Amelio L, Amelio A (2019) Classification methods in image analysis with a special focus on medical analytics, vol 149. Springer, New York. https://doi.org/10.1007/978-3-319-94030-4_3
Qin C, Yao D, Shi Y, Song Z (2018) Computer-aided detection in chest radiography based on artificial intelligence: a survey. Biomed Eng Online 17(1):113. https://doi.org/10.1186/s12938-018-0544-y
Lodwick GS, Keats TE, Dorst JP (1963) The coding of Roentgen images for computer analysis as applied to lung cancer. Radiology 81(2):185–200. https://doi.org/10.1148/81.2.185
Liu Y et al (2012) Computer aided diagnosis system for breast cancer based on color doppler flow imaging. J Med Syst 36(6):3975–3982. https://doi.org/10.1007/s10916-012-9869-4
Diao X-F, Zhang X-Y, Wang T-F, Chen S-P, Yang Y, Zhong L (2011) Highly sensitive computer aided diagnosis system for breast tumor based on color doppler flow images. J Med Syst 35(5):801–809. https://doi.org/10.1007/s10916-010-9461-8
Kaur G, Rana PS, Arora V (2022) State-of-the-art techniques using pre-operative brain MRI scans for survival prediction of glioblastoma multiforme patients and future research directions. Clin Transl Imaging. https://doi.org/10.1007/s40336-022-00487-8
Deng L, Yu D (2014) Deep learning: methods and applications. Found Trends Signal Process 7(3–4):197–387. https://doi.org/10.1561/2000000039
Lladó X et al (2012) Automated detection of multiple sclerosis lesions in serial brain MRI. Neuroradiology 54(8):787–807. https://doi.org/10.1007/s00234-011-0992-6
He Y, Nazir S, Nie B, Khan S, Zhang J (2020) Developing an efficient deep learning-based trusted model for pervasive computing using an LSTM-based classification model. Complexity. https://doi.org/10.1155/2020/4579495
Huo Y, eui Yoon S (2021) A survey on deep learning-based Monte Carlo denoising. Comput Vis Med 7(2):169–185. https://doi.org/10.1007/s41095-021-0209-9
Xiang J, Xu G, Ma C, Hou J (2021) End-to-end learning deep CRF models for multi-object tracking deep CRF models. IEEE Trans Cir and Sys for Video Technol 31(1):275–288. https://doi.org/10.1109/TCSVT.2020.2975842
Yallop A, Seraphin H (2020) Big data and analytics in tourism and hospitality: opportunities and risks. J Tourism Futures 6(3):257–262. https://doi.org/10.1108/JTF-10-2019-0108
Xin M, Wang Y (2019) Research on image classification model based on deep convolution neural network. EURASIP J Image Video Process. https://doi.org/10.1186/s13640-019-0417-8
Das A, Patra GR, Mohanty MN (2020) LSTM based Odia handwritten numeral recognition. In: 2020 International conference on communication and signal processing (ICCSP), pp 538–541. https://doi.org/10.1109/ICCSP48568.2020.9182218
Basheer S, Bhatia S, Sakri SB (2021) Computational modeling of dementia prediction using deep neural network: analysis on OASIS dataset. IEEE Access 9:42449–42462. https://doi.org/10.1109/ACCESS.2021.3066213
O’Shea K, Nash R (2015) An introduction to convolutional neural networks, pp 1–11
Abbas M, Narayan J, Banerjee S, Dwivedy SK (2020) AlexNet based real-time detection and segregation of household objects using Scorbot. In: 4th international conference on computational intelligence and networks, CINE 2020. https://doi.org/10.1109/CINE48825.2020.234392
Abdulsalam Hamwi W, Almustafa MM (2022) Development and integration of VGG and dense transfer-learning systems supported with diverse lung images for discovery of the Coronavirus identity. Inform Med Unlocked 32:101004. https://doi.org/10.1016/j.imu.2022.101004
Akhand MAH, Roy S, Siddique N, Kamal MAS, Shimamura T (2021) Facial emotion recognition using transfer learning in the deep CNN. Electronics 10(9):1036. https://doi.org/10.3390/electronics10091036
Luan S, Chen C, Zhang B, Han J, Liu J (2018) Gabor convolutional networks. IEEE Trans Image Process 27(9):4357–4366. https://doi.org/10.1109/TIP.2018.2835143
Abdallah SE, Elmessery WM, Shams MY, Al-Sattary NSA, Abohany AA, Thabet M (2023) Deep learning model based on ResNet-50 for beef quality classification. Inf Sci Lett 12(1):289–297. https://doi.org/10.18576/isl/120124
Sivaramakrishnan A, Karnan M, Sivakumar R (2014) Medical image analysis—a review. Int J Comput Sci Inf Technol 5(1):236–246
Liu W, Wang Z, Liu X, Zeng N, Liu Y, Alsaadi FE (2017) A survey of deep neural network architectures and their applications. Neurocomputing 234:11–26. https://doi.org/10.1016/j.neucom.2016.12.038
Reema Matthew A, Prasad A, Babu Anto P (2017) A review on feature extraction techniques for tumor detection and classification from brain MRI. In: 2017 international conference on intelligent computing, instrumentation and control technologies, ICICICT 2017, vol 2018, pp 1766–1771. https://doi.org/10.1109/ICICICT1.2017.8342838
Chandrashekar L, Sreedevi A (2017) Assessment of non-linear filters for MRI images. In: Proceedings of the 2017 2nd IEEE international conference on electrical, computer and communication technologies, ICECCT 2017, vol X, no Ii, pp 60–64. https://doi.org/10.1109/ICECCT.2017.8117852
Amiri Golilarz N, Gao H, Kumar R, Ali L, Fu Y, Li C (2020) Adaptive wavelet based MRI brain image de-noising. Front Neurosci 14:728. https://doi.org/10.3389/fnins.2020.00728
Pham C-H et al (2019) Multiscale brain MRI super-resolution using deep 3D convolutional networks. Comput Med Imaging Graphics 77:101647. https://doi.org/10.1016/j.compmedimag.2019.101647
Ito I (2020) A new pseudo-spectral method using the discrete cosine transform. J Imaging 6(4):15. https://doi.org/10.3390/jimaging6040015
Kanoun B, Ambrosanio M, Baselice F, Ferraioli G, Pascazio V, Gómez L (2020) Anisotropic weighted KS-NLM filter for noise reduction in MRI. IEEE Access 8:184866–184884. https://doi.org/10.1109/ACCESS.2020.3029297
Subudhi A, Dash M, Sabut S (2020) Automated segmentation and classification of brain stroke using expectation-maximization and random forest classifier. Biocybern Biomed Eng 40(1):277–289. https://doi.org/10.1016/j.bbe.2019.04.004
Borys D, Serafin W, Frackiewicz M, Psiuk-Maksymowicz K, Palus H (2018) A Phantom study of new bias field correction method combining N3 and KHM for MRI imaging. In: 2018 14th international conference on signal-image technology internet-based systems (SITIS), pp 314–319. https://doi.org/10.1109/SITIS.2018.00055
El-Hag NA et al (2021) Utilization of image interpolation and fusion in brain tumor segmentation. Int J Numer Method Biomed Eng 37(8):e3449. https://doi.org/10.1002/cnm.3449
Chaudhari AS et al (2018) Super-resolution musculoskeletal MRI using deep learning. Magn Reson Med 80(5):2139–2154. https://doi.org/10.1002/mrm.27178
Shen D (2007) Image registration by local histogram matching. Pattern Recognit 40(4):1161–1172. https://doi.org/10.1016/j.patcog.2006.08.012
Hadas I et al (2020) Subgenual cingulate connectivity and hippocampal activation are related to MST therapeutic and adverse effects. Transl Psychiatry 10(1):392. https://doi.org/10.1038/s41398-020-01042-7
Panebianco V et al (2018) Multiparametric magnetic resonance imaging for bladder cancer: development of VI-RADS (Vesical Imaging-Reporting and Data System). Eur Urol 74(3):294–306. https://doi.org/10.1016/j.eururo.2018.04.029
Jin C, Ke S-W (2017) Content-based image retrieval based on shape similarity calculation. 3D Res 8(3):23. https://doi.org/10.1007/s13319-017-0132-0
Ganeshan B et al (2021) Texture analysis of fractional water content images acquired during PET/MRI: initial evidence for an association with total lesion glycolysis survival and gene mutation profile in primary colorectal cancer. Cancers 13(11):2715. https://doi.org/10.3390/cancers13112715
Billings JCW, Thompson GJ, Pan W-J, Magnuson ME, Medda A, Keilholz S (2018) Disentangling multispectral functional connectivity with wavelets. Front Neurosci 12:812. https://doi.org/10.3389/fnins.2018.00812
Lin J-M (2018) Python Non-Uniform Fast Fourier Transform (PyNUFFT): an accelerated non-cartesian MRI package on a heterogeneous platform (CPU/GPU). J Imaging 4(3):51. https://doi.org/10.3390/jimaging4030051
Muzik O, Chugani DC, Juhász C, Shen C, Chugani HT (2000) Statistical parametric mapping: assessment of application in children. Neuroimage 12(5):538–549. https://doi.org/10.1006/nimg.2000.0651
Theaud G, Houde J-C, Boré A, Rheault F, Morency F, Descoteaux M (2020) TractoFlow: a robust, efficient and reproducible diffusion MRI pipeline leveraging Nextflow & Singularity. Neuroimage 218:116889. https://doi.org/10.1016/j.neuroimage.2020.116889
Kurup RV, Sowmya V, Soman KP (2020) ICICCT 2019—system reliability, quality control, safety, maintenance and management. Springer, Singapore. https://doi.org/10.1007/978-981-13-8461-5
Meera R, Anandhan P (2018) A review on automatic detection of brain tumor using computer aided diagnosis system through MRI. EAI Endors Trans Energy Web 5(20):1–10. https://doi.org/10.4108/eai.12-9-2018.155747
Atazandi GR (2019) Automatic optimal thresholding using generalized fuzzy entropies and genetic algorithm. Artif Intell Neurosci 10(2):143–150
Ibrahim RW, Hasan AM, Jalab HA (2018) A new deformable model based on fractional Wright energy function for tumor segmentation of volumetric brain MRI scans. Comput Methods Prog Biomed 163:21–28. https://doi.org/10.1016/j.cmpb.2018.05.031
Bahadure NB, Ray AK, Thethi HP (2018) Comparative approach of MRI-based brain tumor segmentation and classification using genetic algorithm. J Digit Imaging 31(4):477–489. https://doi.org/10.1007/s10278-018-0050-6
Shehata M et al (2018) 3D kidney segmentation from abdominal diffusion MRI using an appearance-guided deformable boundary. PLoS ONE 13(7):1–21. https://doi.org/10.1371/journal.pone.0200082
Agarwal S, Singh OP, Nagaria D (2017) Analysis and comparison of wavelet transforms for denoising MRI image. Biomed Pharmacol J 10(2):831–836. https://doi.org/10.13005/bpj/1174
Sairanen V, Leemans A, Tax CMW (2018) Fast and accurate Slicewise OutLIer Detection (SOLID) with informed model estimation for diffusion MRI data. Neuroimage 181:331–346. https://doi.org/10.1016/j.neuroimage.2018.07.003
Pan Z, Lu J (2007) A bayes-based region-growing algorithm for medical image segmentation. Comput Sci Eng 9(4):32–38. https://doi.org/10.1109/MCSE.2007.67
Mahalakshmi DM, Sumathi S (2019) Brain tumour segmentation strategies utilizing mean shift clustering and content based active contour segmentation. ICTACT J Image Video Process 9(4):2002–2008. https://doi.org/10.21917/ijivp.2019.0284
Sohaib M, Kim JM (2017) Quantitative assessment of heart function: A hybrid mechanism for left ventricle segmentation from cine MRI sequences. In: Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), vol 10142. LNAI, pp 169–179. https://doi.org/10.1007/978-3-319-51691-2_15
Boyes RG et al (2008) Intensity non-uniformity correction using N3 on 3-T scanners with multichannel phased array coils. Neuroimage 39(4):1752–1762. https://doi.org/10.1016/j.neuroimage.2007.10.026
Park J, Lee J, Lee J, Lee SK, Park JY (2020) Strategies for rapid reconstruction in 3D MRI with radial data acquisition: 3D fast Fourier transform vs two-step 2D filtered back-projection. Sci Rep 10(1):1–11. https://doi.org/10.1038/s41598-020-70698-4
Tabelow K et al (2019) hMRI—a toolbox for quantitative MRI in neuroscience and clinical research. Neuroimage 194:191–210. https://doi.org/10.1016/j.neuroimage.2019.01.029
Lin W et al (2018) Convolutional neural networks-based MRI image analysis for the Alzheimer’s disease prediction from mild cognitive impairment. Front Neurosci 12:1–13. https://doi.org/10.3389/fnins.2018.00777
Tibrewala R, Pedoia V, Bucknor M, Majumdar S (2020) Principal component analysis of simultaneous PET-MRI reveals patterns of bone-cartilage interactions in osteoarthritis. J Magn Reson Imaging 52(5):1462–1474. https://doi.org/10.1002/jmri.27146
Zhang L, Ji Q (2011) A bayesian network model for automatic and interactive image segmentation. IEEE Trans Image Process 20(9):2582–2593. https://doi.org/10.1109/TIP.2011.2121080
Zhuang J, Yang J, Gu L, Dvornek N (2019) Shelfnet for fast semantic segmentation. In: Proceedings—2019 international conference on computer vision workshop, ICCVW 2019, pp 847–856. https://doi.org/10.1109/ICCVW.2019.00113
Chen L-C, Papandreou G, Kokkinos I, Murphy K, Yuille AL (2018) DeepLab: semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. IEEE Trans Pattern Anal Mach Intell 40(4):834–848. https://doi.org/10.1109/TPAMI.2017.2699184
Ronneberger O, Fischer P, Brox T (2015) U-Net: convolutional networks for biomedical image segmentation BT. In: Medical image computing and computer-assisted intervention—MICCAI 2015, pp 234–241
Lateef F, Ruichek Y (2019) Survey on semantic segmentation using deep learning techniques. Neurocomputing 338:321–348. https://doi.org/10.1016/j.neucom.2019.02.003
Li B, Liu S, Xu W, Qiu W (2018) Real-time object detection and semantic segmentation for autonomous driving. In: Proc. of SPIE, vol 10608. https://doi.org/10.1117/12.2288713
Zhang Y, Chen H, He Y, Ye M, Cai X, Zhang D (2018) Road segmentation for all-day outdoor robot navigation. Neurocomputing 314:316–325. https://doi.org/10.1016/j.neucom.2018.06.059
Tao X, Zhang D, Ma W, Liu X, Xu D (2018) Automatic metallic surface defect detection and recognition with convolutional neural networks. Appl Sci 8(9):1575. https://doi.org/10.3390/app8091575
Kemker R, Salvaggio C, Kanan C (2018) Algorithms for semantic segmentation of multispectral remote sensing imagery using deep learning. ISPRS J Photogramm Remote Sens 145:60–77. https://doi.org/10.1016/j.isprsjprs.2018.04.014
Ji Y, Zhang H, Jonathan Wu QM (2018) Salient object detection via multi-scale attention CNN. Neurocomputing 322:130–140. https://doi.org/10.1016/j.neucom.2018.09.061
Krasovskaya S, MacInnes WJ (2019) Salience models: a computational cognitive neuroscience review. Vision 3(4):56. https://doi.org/10.3390/vision3040056
Milioto A, Lottes P, Stachniss C (2018) Real-time semantic segmentation of crop and weed for precision agriculture robots leveraging background knowledge in CNNs. In: 2018 IEEE international conference on robotics and automation (ICRA), pp 2229–2235.https://doi.org/10.1109/ICRA.2018.8460962
Hafiz AM, Bhat GM (2020) A survey on instance segmentation: state of the art. Int J Multimed Inf Retr 9(3):171–189. https://doi.org/10.1007/s13735-020-00195-x
Hunter P (2019) The advent of AI and deep learning in diagnostics and imaging. EMBO Rep 20(7):e48559. https://doi.org/10.15252/embr.201948559
Kiranyaz S, Ince T, Abdeljaber O, Avci O, Gabbouj M (2019) 1-D convolutional neural networks for signal processing applications. In: ICASSP 2019—2019 IEEE international conference on acoustics, speech and signal processing (ICASSP), pp 8360–8364. https://doi.org/10.1109/ICASSP.2019.8682194
Brinker TJ et al (2018) Skin cancer classification using convolutional neural networks: systematic review. J Med Internet Res 20(10):1–8. https://doi.org/10.2196/11936
He K, Gkioxari G, Dollár P, Girshick R (2020) Mask R-CNN. IEEE Trans Pattern Anal Mach Intell 42(2):386–397. https://doi.org/10.1109/TPAMI.2018.2844175
Li Y, Qi H, Dai J, Ji X, Wei Y (2017) Fully convolutional instance-aware semantic segmentation. In: Proceedings—30th IEEE conference on computer vision and pattern recognition, CVPR 2017, vol 2017, pp 4438–4446. https://doi.org/10.1109/CVPR.2017.472
Wang P et al (2018) Understanding convolution for semantic segmentation. In: 2018 IEEE winter conference on applications of computer vision (WACV), pp 1451–1460.https://doi.org/10.1109/WACV.2018.00163
Brünger J, Gentz M, Traulsen I, Koch R (2020) Panoptic segmentation of individual pigs for posture recognition. Sensors 20(13):1–21. https://doi.org/10.3390/s20133710
Chen H, Qi X, Yu L, Heng PA (2016) DCAN: deep contour-aware networks for accurate gland segmentation. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit 2016:2487–2496. https://doi.org/10.1109/CVPR.2016.273
Zeng C, Gu L, Liu Z, Zhao S (2020) Review of deep learning approaches for the segmentation of multiple sclerosis lesions on brain MRI. Front Neuroinform 14:1–8. https://doi.org/10.3389/fninf.2020.610967
Zhang L et al (2020) Block level skip connections across cascaded V-net for multi-organ segmentation. IEEE Trans Med Imaging 39(9):2782–2793. https://doi.org/10.1109/TMI.2020.2975347
Nikan S et al (2021) PWD-3DNet: a deep learning-based fully-automated segmentation of multiple structures on temporal bone CT scans. IEEE Trans Image Process 30:739–753. https://doi.org/10.1109/TIP.2020.3038363
Henry T et al (2021) Brain tumor segmentation with self-ensembled, deeply-supervised 3D U-net neural networks: a BraTS 2020 challenge solution, pp 327–339. https://doi.org/10.1007/978-3-030-72084-1_30
Mahmoudi L, El Zaart A (2012) A survey of entropy image thresholding techniques. In: 2012 2nd international conference on advances in computational tools for engineering applications (ACTEA), pp 204–209. https://doi.org/10.1109/ICTEA.2012.6462867
Chakraborty R, Sushil R, Garg ML (2019) An improved PSO-based multilevel image segmentation technique using minimum cross-entropy thresholding. Arab J Sci Eng 44(4):3005–3020. https://doi.org/10.1007/s13369-018-3400-2
Wang H, Jiang Y, Jiang X, Wu J, Yang X (2018) Automatic vessel segmentation on fundus images using vessel filtering and fuzzy entropy. Soft Comput 22(5):1501–1509. https://doi.org/10.1007/s00500-017-2872-4
Wunnava A, KumarNaik M, Panda R, Jena B, Abraham A (2020) A differential evolutionary adaptive Harris hawks optimization for two dimensional practical Masi entropy-based multilevel image thresholding. J King Saud Univ Comput Inf Sci. https://doi.org/10.1016/j.jksuci.2020.05.001
Roche A, Ribes D, Bach-Cuadra M, Krüger G (2011) On the convergence of EM-like algorithms for image segmentation using Markov random fields. Med Image Anal 15(6):830–839. https://doi.org/10.1016/j.media.2011.05.002
Raja NSM, Fernandes SL, Dey N, Satapathy SC, Rajinikanth V (2018) Contrast enhanced medical MRI evaluation using Tsallis entropy and region growing segmentation. J Ambient Intell Humaniz Comput. https://doi.org/10.1007/s12652-018-0854-8
Wang W, Chen J, Cao J (2019) Using low-field NMR and MRI to characterize water status and distribution in modified wood during water absorption. Holzforschung 73(11):997–1004. https://doi.org/10.1515/hf-2018-0293
Ammar A, Bouattane O, Youssfi M (2019) Review and comparative study of three local based active contours optimizers for image segmentation. In: 2019 5th international conference on optimization and applications (ICOA), pp 1–6. https://doi.org/10.1109/ICOA.2019.8727683
Nilakant R, Menon HP, Vikram K (2017) A survey on advanced segmentation techniques for brain MRI.pdf, vol 7, no 4, pp 1448–1456
Wu J, Zhang Y, Tang X (2019) A multi-atlas guided 3D fully convolutional network for mri-based subcortical segmentation. In: 2019 IEEE 16th international symposium on biomedical imaging (ISBI 2019), pp 705–708. https://doi.org/10.1109/ISBI.2019.8759286
Sun L, Shao W, Wang M, Zhang D, Liu M (2020) High-order feature learning for multi-atlas based label fusion: application to brain segmentation with MRI. IEEE Trans Image Process 29:2702–2713. https://doi.org/10.1109/TIP.2019.2952079
Alang TAIT, Tan TS, Yaakub A (2020) Implementation of circular Hough transform on MRI images for eye globe volume estimation. Int J Biomed Eng Technol 33(2):123–133. https://doi.org/10.1504/IJBET.2020.107708
Oliveira DA, Silva MET, Pouca MV, Parente MPL, Mascarenhas T, Natal Jorge RM (2020) Biomechanical simulation of vaginal childbirth: the colors of the pelvic floor muscles. https://doi.org/10.1007/978-3-030-15923-8_1
Pham DD, Morariu CA, Terheiden T, Landgraeber S, Jäger M, Pauli J (2017) Mri hip joint segmentation: a locally bhattacharyya weighted hybrid 3d level set approach. In: VCBM 2017—Eurographics workshop on visual computing for biology and medicine, pp 113–117. https://doi.org/10.2312/vcbm.20171243
Colliot O, Camara O, Bloch I (2006) Integration of fuzzy spatial relations in deformable models-application to brain MRI segmentation. Pattern Recognit 39(8):1401–1414. https://doi.org/10.1016/j.patcog.2006.02.022
Huang Z, Wang X, Wang J, Liu W, Wang J (2018) Weakly-Supervised semantic segmentation network with deep seeded region growing. In: Proceedings of the IEEE computer society conference on computer vision and pattern recognition, pp 7014–7023. https://doi.org/10.1109/CVPR.2018.00733
Katoch S, Chauhan SS, Kumar V (2021) A review on genetic algorithm: past, present, and future. Multimed Tools Appl 80(5):8091–8126. https://doi.org/10.1007/s11042-020-10139-6
Ji DX, Foong KWC, Ong SH (2013) A two-stage rule-constrained seedless region growing approach for mandibular body segmentation in MRI. Int J Comput Assist Radiol Surg 8(5):723–732. https://doi.org/10.1007/s11548-012-0806-2
Ali RA, Abood LK (2017) Automatic brain tumor segmentation from MRI Images using superpixels based split and Merge algorithm. Int J Sci Res 6(7):274–278. https://doi.org/10.21275/art20174940
Kim HH, Kim Y, Park YR (2021) Interpretable conditional recurrent neural network for weight change prediction: algorithm development and validation study. JMIR Mhealth Uhealth 9(3):e22183. https://doi.org/10.2196/22183
Esteva A et al (2017) Dermatologist-level classification of skin cancer with deep neural networks. Nature 542(7639):115–118. https://doi.org/10.1038/nature21056
Lakhani P, Sundaram B (2017) Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology 284(2):574–582. https://doi.org/10.1148/radiol.2017162326
Park SH, Han K (2018) Methodologic guide for evaluating clinical performance and effect of artificial intelligence technology for medical diagnosis and prediction. Radiology 286(3):800–809. https://doi.org/10.1148/radiol.2017171920
Zhang W et al (2015) Deep convolutional neural networks for multi-modality isointense infant brain image segmentation. Neuroimage 108:214–224. https://doi.org/10.1016/j.neuroimage.2014.12.061
Chen H, Dou Q, Yu L, Qin J, Heng P-A (2018) VoxResNet: deep voxelwise residual networks for brain segmentation from 3D MR images. Neuroimage 170:446–455. https://doi.org/10.1016/j.neuroimage.2017.04.041
Zikic D, Ioannou Y, Criminisi A, Brown M (2014) Segmentation of brain tumor tissues with convolutional neural networks. In: MICCAI workshop on multimodal brain tumor segmentation challenge (BRATS), MICCAI wor
Urban G, Bendszus M, Hamprecht FA, Kleesiek J (2014) Multi-modal brain tumor segmentation using deep convolutional neuralnetworks. In: MICCAI BraTS (Brain Tumor Segmentation) challenge. proceedings, winningcontribution, pp 31–35
Dvořák P, Menze B (2016) Local structure prediction with convolutional neural networks for multimodal brain tumor segmentation. In: Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), vol 9601 LNCS, pp 59–71. https://doi.org/10.1007/978-3-319-42016-5_6
Yogananda CGB et al (2020) A fully automated deep learning network for brain tumor segmentation. Tomography 6(2):186–193. https://doi.org/10.18383/j.tom.2019.00026
Milletari F et al (2017) Hough-CNN: deep learning for segmentation of deep brain regions in MRI and ultrasound. Comput Vis Image Underst 164:92–102. https://doi.org/10.1016/j.cviu.2017.04.002
Bernal J et al (2019) Deep convolutional neural networks for brain image analysis on magnetic resonance imaging: a review. Artif Intell Med 95:64–81. https://doi.org/10.1016/j.artmed.2018.08.008
Sajid S, Hussain S, Sarwar A (2019) Brain tumor detection and segmentation in MR images using deep learning. Arab J Sci Eng 44(11):9249–9261. https://doi.org/10.1007/s13369-019-03967-8
Zhao L, Jia K (2016) Multiscale CNNs for brain tumor segmentation and diagnosis. Comput Math Methods Med 2016:8356294. https://doi.org/10.1155/2016/8356294
Havaei M et al (2017) Brain tumor segmentation with Deep Neural Networks. Med Image Anal 35:18–31. https://doi.org/10.1016/j.media.2016.05.004
Ather D et al (2022) Selection of smart manure composition for smart farming using artificial intelligence technique. J Food Qual 2022:1–7. https://doi.org/10.1155/2022/4351825
Liu S, Liu G, Zhou H (2019) A robust parallel object tracking method for illumination variations. Mob Netw Appl 24(1):5–17. https://doi.org/10.1007/s11036-018-1134-8
Liu S, Liu X, Wang S, Muhammad K (2021) Fuzzy-aided solution for out-of-view challenge in visual tracking under IoT-assisted complex environment. Neural Comput Appl 33(4):1055–1065. https://doi.org/10.1007/s00521-020-05021-3
Huang C et al (2020) Sample imbalance disease classification model based on association rule feature selection. Pattern Recognit Lett 133:280–286. https://doi.org/10.1016/j.patrec.2020.03.016
Brosch T, Tang LYW, Yoo Y, Li DKB, Traboulsee A, Tam R (2016) Deep 3D convolutional encoder networks with shortcuts for multiscale feature integration applied to multiple sclerosis lesion segmentation. IEEE Trans Med Imaging 35(5):1229–1239. https://doi.org/10.1109/TMI.2016.2528821
Wang H, Zhang Z, Han S (2021) SpAtten: efficient sparse attention architecture with cascade token and head pruning. In: 2021 IEEE international symposium on high-performance computer architecture (HPCA), pp 97–110.https://doi.org/10.1109/HPCA51647.2021.00018
Nasor M, Obaid W (2020) Detection and localization of early-stage multiple brain tumors using a hybrid technique of patch-based processing, K-means clustering and object counting. Int J Biomed Imaging 2020:9035096. https://doi.org/10.1155/2020/9035096
Hu A et al (2020) A parallel and cascade control system: Magnetofection of miR125b for synergistic tumor-Association macrophage polarization regulation and tumor cell suppression in breast cancer treatment. Nanoscale 12(44):22615–22627. https://doi.org/10.1039/d0nr06060g
Maleki M, Teshnehlab PM, Nabavi M (2012) Diagnosis of Multiple Sclerosis (MS) Using Convolutional Neural Network (CNN) from MRIs. Glob J Med Plant Res 1(1):50–54
Cui S, Mao L, Jiang J, Liu C, Xiong S (2018) Automatic semantic segmentation of brain gliomas from MRI images using a deep cascaded neural network. J Healthc Eng. https://doi.org/10.1155/2018/4940593
Kleesiek J et al (2016) Deep MRI brain extraction: a 3D convolutional neural network for skull stripping. Neuroimage 129:460–469. https://doi.org/10.1016/j.neuroimage.2016.01.024
Li W, Wang G, Fidon L, Ourselin S, Cardoso MJ, Vercauteren T (2017) On the compactness, efficiency, and representation of 3D convolutional networks: brain parcellation as a pretext task. In: Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), vol 10265 LNCS, pp 348–360. https://doi.org/10.1007/978-3-319-59050-9_28
Wachinger C, Reuter M, Klein T (2018) DeepNAT: deep convolutional neural network for segmenting neuroanatomy. Neuroimage 170:434–445
Paulsen RR, Pedersen KS (2015) Image analysis: 19th Scandinavian conference, SCIA 2015 Copenhagen, Denmark, June 15–17, 2015 proceedings. In: Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), vol 9127, pp 201–211. https://doi.org/10.1007/978-3-319-19665-7
Vaidya S, Chunduru A, Muthuganapathy R, Krishnamurthi G (2015) Longitudinal multiple sclerosis lesion segmentation using 3d convolutional neural networks. In: Vaidya S, Chunduru A, Muthuganapathy R, Krishnamurthi G (eds) Biomedical Imaging Lab. Department of Engineering Design Indian Institute
Valverde S et al (2017) Improving automated multiple sclerosis lesion segmentation with a cascaded 3D convolutional neural network approach. Neuroimage 155:159–168. https://doi.org/10.1016/j.neuroimage.2017.04.034
Ding Y et al (2020) Using deep convolutional neural networks for neonatal brain image segmentation. Front Neurosci 14:207. https://doi.org/10.3389/fnins.2020.00207
Chandrakar MK, Mishra A (2020) Brain tumor detection using multipath Convolution Neural Network (CNN). Int J Comput Vis Image Process 10(4):43–53. https://doi.org/10.4018/IJCVIP.2020100103
Ben naceur M, Saouli R, Akil M, Kachouri R (2018) Fully automatic brain tumor segmentation using end-to-end incremental deep neural networks in MRI images. Comput Methods Prog Biomed 166:39–49. https://doi.org/10.1016/j.cmpb.2018.09.007
Moeskops P et al (2018) Evaluation of a deep learning approach for the segmentation of brain tissues and white matter hyperintensities of presumed vascular origin in MRI. Neuroimage Clin 17:251–262. https://doi.org/10.1016/j.nicl.2017.10.007
Sudre CH, Li W, Vercauteren T, Ourselin S, Jorge Cardoso M (2017) Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations. In: Deep learning in medical image analysis and multimodal learning for clinical decision support: Third International Workshop, DLMIA 2017, and 7th International Workshop, ML-CDS 2017, held in conjunction with MICCAI 2017 Quebec City, vol 2017, pp 240—248. https://doi.org/10.1007/978-3-319-67558-9_28
Khalifa NE, Loey M, Mirjalili S (2021) A comprehensive survey of recent trends in deep learning for digital images augmentation. Artif Intell Rev. https://doi.org/10.1007/s10462-021-10066-4
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors state that they have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kshatri, S.S., Singh, D. Convolutional Neural Network in Medical Image Analysis: A Review. Arch Computat Methods Eng 30, 2793–2810 (2023). https://doi.org/10.1007/s11831-023-09898-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11831-023-09898-w