Abstract
Severe winter wheat yield losses due to infestations of wheat curl mite, Aceria tosichella Keifer, and mite-transmitted viruses occur in wheat production areas of the United States and Canada. Mite infestation alone causes stunted, chlorotic plants in susceptible wheat varieties, and mites transmit Wheat Streak Mosaic (WSMV), High Plains Wheat Mosaic (HPWMoV), and Triticum Mosaic Virus (TriMV). Wheat curl mites were collected from 25 sites in Kansas, Missouri, Nebraska, Texas, North Dakota, and South Dakota in 2014 and 2015. At each site, mite virulence was determined to wheat plants harboring the Cmc2-, Cmc3-, or Cmc4 mite resistance gene; or Cmc4 plus the Wsm2 WSMV resistance gene. Mites collected from 92%, 36%, and 24% of sites were virulent to susceptible Jagger wheat plants (no Cmc), Cmc2, and Cmc3, respectively. The mega-population consisting of all 25 mite sub-populations was avirulent to 80% of plants containing Cmc4 + Wsm2 or Cmc4. WSMV, HPWMoV, or TriMV was present in mites at 76%, 16%, and 8% of the 25 sites, respectively. Our results will enable breeders to increase the efficiency of wheat production by releasing wheat varieties containing wheat curl mite resistance genes that reduce wheat yield losses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The wheat curl mite, Aceria tosichella Keifer, an important arthropod pest of winter wheat, Triticum aestivum L., is the only known vector of several viruses that significantly reduce wheat yields. A. tosichella transmits Wheat streak mosaic virus (WSMV, family Potyviridae, genus Tritimovirus) (Orlob 1966; Slykhuis 1955); High Plains wheat mosaic virus (HPWMoV, genus Emaravirus, formerly High Plains virus; www.ictvonline.org/proposals-15/2015.018aP.A.v3.Emaravirus_sp.pdf) (Seifers et al. 1997); and Triticum Mosaic Virus (TriMV, family Potyviridae, genus Poacevirus) (Seifers et al. 2008, 2009b).
Yield losses from infection by WSMV, the most prominent of the three viruses, have averaged from 2.5 to 5% over the past 25 years (Christian and Willis 1993; French and Stenger 2003) depending on wheat cultivar (Martin et al. 1984). However, past outbreak WSMV infections in Canada and Kansas demonstrate that yield reductions of > 10% can occur (Atkinson and Grant 1967; Sim et al. 1988). Aggregate yield losses from single or multiple viral coinfections are prevalent in the central United States wheat production area (Mahmood et al. 1998; Byamukama et al. 2016).
Effective A. tosichella population control is difficult for several reasons. While prediction of infestations is possible if pre-harvest volunteer wheat or green corn are near winter wheat emerging in the fall, such predictions are complicated because many species of range grasses are mite hosts (Slykhuis 1955; Connin 1956; Skoracka et al. 2012). Delaying wheat planting until volunteer wheat is removed also is problematic because the uniformity of volunteer wheat destruction by producers varies widely. In reality, planting dates are normally determined by availability of soil moisture. In addition, delayed planting is not feasible for growers using wheat for winter livestock forage (Martin et al. 1984; Velandia et al. 2010). Finally, no effective acaricides exist for A. tosichella management (Townsend and Johnson 1996; Morgan et al. 2005; McMechan and Hein 2016).
Thus, wheat plant resistance is likely to be the most viable, economical, and environmentally-safe and eco-friendly approach to wheat curl mite population reduction (Smith 1999). Andrews and Slykhuis (1956) were the first to identify mite resistance in tall wheatgrass, Agropyron elongatum (Host.) P. Beaur., and Agropyron intermedium (Host.) P. Beaur. Harvey and Livers (1975) determined that genes from rye, Secale cereale L., suppress mite populations more than genes from wheat, and Martin et al. (1976) showed that Salmon, a wheat cultivar carrying a segment of rye chromosome 1R, was highly resistant to wheat curl mite but susceptible to WSMV. Soon after, Martin et al. (1984) highlighted that resistance to wheat curl mite in Salmon reduced the occurrence of WSMV by 58%. Harvey and Martin (1980) and Harvey et al. (1990) found that cultivars with increased trichome density had reduced wheat curl mite populations. Numerous wheat cultivars with genetic resistance to mites have been used successfully to suppress mite populations and lower the level of infection of mite-vectored viruses by inhibiting mite reproductive capacity (Harvey and Martin 1988; Harvey et al. 1994, 2005; Conner et al. 1991).
Four cereal genes that express resistance to wheat curl mite have been deployed in commercial varieties in North America. The gene Cmc1 from Tausch’s goatgrass, Aegilops tauschii (Coss.). Schmal. (syn. Ae. squarrosa L.; Triticum tauschii) was introgressed to wheat chromosome 6D (Thomas and Conner 1986; Whelan and Thomas 1989); in wheat cultivar Radiant (Thomas et al. 2012). The gene Cmc4 in wheat cultivars MT06X424 (Hofer et al. 2011) and OK05312 (Carver et al. 2016), also from Ae. tauschii, was shown to segregate independently of Cmc1 on wheat chromosome 6D by Malik et al. (2003). The gene Cmc2 (PI52452) originated from A. elongatum and was identified by Martin et al. (1976) and Whelan and Hart (1988); and Cmc3, the rye gene in wheat cultivars Salmon and TAM107 (Martin et al. 1983; Schlegel and Kynast 1987), was mapped and named by Malik et al. (2003). The Wsm1, Wsm2, and Wsm3 WSMV resistance genes have been identified (Triebe et al. 1991; Liu et al. 2011; Lu et al. 2011), but no HPWMoV resistance genes have been identified. Currently, no known commercial wheat varieties contain combined resistance to wheat curl mite and WSMV (Graybosch et al. 2009; DeWolf et al. 2014).
The term virulence in studies of plant resistance to arthropods is defined as the ability of an arthropod to overcome a plant resistance gene (Smith 2005; Kobayashi 2016; O’Neal et al. 2018) or conversely, the loss of a resistant plant’s capability to distinguish the presence of the arthropod, due to mutation in an arthropod avirulence gene(s) (Smith and Clement 2012). The limitation of using any Cmc gene is that virulent wheat curl mite populations may preexist or develop to overcome resistance (Harvey et al. 1995, 1997, 1999). Thus, new sources of mite resistance must be available to wheat breeding programs to manage wheat curl mite virulence effectively.
Although mite virulence patterns in North America have not been examined since 1998, the fact that no major mite outbreak infestations have occurred indicates that mite populations have remained avirulent to Cmc4 (Cmc4 has remained resistant), and that mite virulence to Cmc2 and Cmc3 (Cmc2 and Cmc3 susceptibility) reported by Harvey et al. (1995, 1999) remains. However, studies of Cmc4 field resistance have been limited to Oklahoma and Montana (Carver et al. 2016; Hofer et al. 2011). Therefore, we hypothesized that mite virulence responses to Cmc2, Cmc3, and Cmc4 may have changed in locations other than these in the U.S. Great Plains and further, that virulence may be related to the distribution of mite-transmitted viruses. The goals of this study were to assess the virulence status of mite populations in wheat from 25 different geographic locations in the U.S. Great Plains to mite resistance genes in wheat; and to determine the incidence of WSMV, HPWMoV, and TriMV in mites collected from each location.
Materials and methods
Collection of A. toschiella populations
Wheat curl mites were collected from grain heads of wheat, Triticum aestivum L., at 25 locations in the U.S. Great Plains from May 21 to July 10, 2014 and June 25 to July 12, 2015 (Supplemental Table 1). The selection of locations was based on results of Harvey et al. (1995, 1997, 1999) and on reference data describing areas of high U.S. wheat production (USDA NASS 2013). Thirty wheat heads were sampled randomly from each of three fields at each location (distance between fields was ~ 500 m). The resulting total of 90 heads per location provided an adequate representation of the mite population at each location. Ten wheat heads, each containing ~ 1000 mites/head, were selected at random from the 90 heads collected at each location and used to establish a mite colony for each population. 100 mites from heads at each location were placed separately on each of 100 susceptible Jagger plants in 45 × 45 × 75 cm cages (BioQuip, Rancho Dominguez, CA, USA) covered with 35-micron mite-proof mesh (Elko Filtering Co., Miami, FL USA; Item # 03-35/16). In all experiments, Sungrow METRO-MIX 360 soil was used (Hummert International, Topeka, KS, USA) to grow wheat varieties and planted one seed per pot. Each colony was checked 21 days post-establishment under the microscope to make ensure successful mite transfers. An 8 h recess was observed between transfers with each population to prevent cross-contamination of colonies (Orlob 1966). After colonies were established, DNA was extracted from mites in each colony and PCR amplified with a nuclear ribosomal internal transcribed spacer 1 (ITS1) using protocols of Malik (2001) to verify that each colony contained only A. tosichella and free of contamination of other Aceria spp.
A. tosichella virulence to mite resistance genes
Experiments were conducted to assess differences in mite population increase and mite-related leaf rolling on plants containing different wheat curl mite resistance genes. All known commercial wheat varieties lack combined resistance to WSMV and wheat curl mite (DeWolf et al. 2014; Graybosch et al. 2009). Three wheat genotypes with different Cmc genes were assessed for wheat curl mite virulence: PI52452, which contains Cmc2; TAM 107, which contains Cmc3; and advanced Kansas breeding line KSU2R-2, containing Cmc4 and Wsm2. The pedigree of KSU2R-2 is RonL/U5287//KS06O3A ~ 58. The cultivars Jagger and OK05312, containing Cmc4 (Carver et al. 2016) were included as susceptible and resistant controls, respectively. Plants of all these wheat genotypes tested in all experiments were maintained in the greenhouse at 24:20 °C day/night and a 14:10 [L:D] h photoperiod.
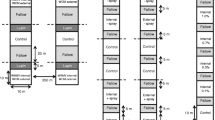
Individual experiments were conducted with each of the 25 mite populations collected from the locations described in Supplemental Table 1. Each experiment contained a total of 25 plants, consisting of five two-leaf-stage seedlings (replications) of each of the three test genotypes, the OK05312 resistant cultivar and the Jagger susceptible control. Each seedling was infested in the second leaf stage with a piece of wheat leaf containing 10 wheat curl mite adults and all 25 plants were placed in a completely randomized design in a 45 × 45 × 75 cm cage (BioQuip, Rancho Dominguez, CA, USA) covered with 35 micron mite-proof mesh (Elko Filtering Co., Miami, FL USA) in a manner that prohibited leaf-to leaf contact (Fig. 1 ). Each cage was rotated 90° southward daily to ensure that each caged plant had an equal opportunity to receive sunlight and airflow. This process was repeated for each of the 25 mite populations.
Flow chart of methodology used to assess mite population growth. Left: 2-leaf seedlings infested with a piece of wheat leaf containing 10 mite females; Center: cage containing 25 plants, consisting of 5 plants (replicates) of each of 3 wheat test genotypes and 2 controls. Right: plant processing for mite population counts
Each caged plant was assessed for mite-induced leaf folding using protocols of Chuang et al. (2017) at 14 days post infestation, and all plants were then cut just above the soil level and their leaves were folded and spread on adhesive 5 × 9 cm gridded blue paper sheets. Each sheet was then stored in a 50 mL Falcon tube (Fisher Scientific, Waltham, MA, USA) for 4–5 days or until the leaves dried at room temperature) and placed in a tube holder at a 45° angle to prevent mites from falling into the bottom of the tube (Murugan et al. 2011). Mites migrated from leaves to the adhesive and were counted using a Nikon SMZ-645 stereo zoom microscope at × 50 magnification.
Comparison of virus infections in A. tosichella populations
To determine the degree of infection in each mite population by WSMV, HPWMoV, or TriMV, 15 two-leaf-stage seedlings of the mite-susceptible cultivar Jagger were each infested in the second leaf stage with a piece of wheat leaf containing 10 mites from each of the 25 populations. Plants were caged for 21 days to allow virus infection symptoms to occur using protocols of Chuang et al. (2017). Each cage represented an individual experiment with each mite population. Five plants from each group of 15 infested seedlings and two negative control plants of the susceptible cultivar Jagger were subjected to ELISA for each virus using protocols of Seifers et al. (2009a, b). Briefly, viruses were detected using Agdia virus-specific ELISA kits SRA 47001/0096, SRA 17200/0096, and RS 21300/0096 for WSMV, HPWMo, and TriMV, respectively. Each virus was assayed by coating reader plate wells with carbonate coating buffer/capture antibody mixture and incubating overnight in a humid box at 4C, according to manufacturer’s instructions. Plant leaf tissue was homogenized in 1:10 (g/v) ratio in PBST buffer and diluted 1:100 in PBST for further processing. Diluted plant extracts were dispensed in 100 ul aliquots/well and each assay was completed as instructed by the manufacturer.
Absorbance values at 405 nm were then measured after 60 min, using a Vmax kinetic microplate reader (Molecular Devices, San Francisco, CA, USA) and used to calculate virus infection rates in plants fed on by mites from each location. Samples were determined to be positive for a virus if they contained 3× the average absorbance values of uninfected Jagger leaves (Fahim et al. 2012; Rotenberg et al. 2016). All ELISA values are shown in Supplemental Table 2.
The rate of infection of each virus from mites in each population was determined using an ELISA ratio calculated as: OD405 nm value of one leaf of an infected plant/OD405 nm value of one leaf of an uninfected control plant. A high ELISA ratio indicates high virus infection and a low ratio indicates comparatively reduced- or no infection. Seifers et al. (2009a) used the ELISA ratio to describe GHVs (Greater than Healthy plant Values), as the number of times greater an ELISA ratio is than the ELISA value of the healthy control. Mite populations infesting plants exhibiting ELISA ratios significantly higher than plants infested by other mite populations were considered to have a significantly greater infection capacity.
Statistical analyses
Data for mite-induced leaf folding, mite numbers. and mite virus infection were analyzed independently for each of the 25 mite populations. Leaf folding data were analyzed by comparing each mite-resistant genotype (Cmc2, Cmc3, Cmc4 + Wsm2) to resistant- and susceptible control cultivars using the χ2 Fisher’s Exact Test (Fisher 1954). If the χ2 test was significant at P < 0.05 for the complete experiment, individual statistical treatment differences were displayed as significant (0.05), highly significant (0.01), or non-significant at 0.05.
Mite numbers and ELISA virus ratio infection data were subjected to one-way ANOVA using PROC GLIMMIX (SAS 2008). These data did not follow assumptions of normality and homogeneity of variances as indicated by the Shapiro–Wilk test of normality and the Brown–Forsythe and Levene tests of homogeneity of variances (Shapiro and Francia 1972). Both mite number data and ELISA virus ratio data were fit to gamma distributions for analysis after log transformation. Mean ± 95% CI numbers of mites and mean ± 95% CI ELISA ratios were separated by Tukey’s HSD (honestly significant difference) test or least significant difference (LSD) test if the type III test for fixed effect was significant at P < 0.05. The LSD test, recommended for exploratory studies by Milliken and Johnson (2004) such as these, was conducted when the Tukey’s HSD test was too conservative. Mite virulence (Fig. 2) was visualized using the computation tool Circos 0.69–4 (Krzywinski et al. 2009) by comparing the mean numbers of mites from each geographic population on each wheat genotype containing the mite resistance genes Cmc2, Cmc3, Cmc4, and Cmc4 plus Wsm2. Color bands represent numbers of mites from each population on plants of each genotype; with narrow bands indicating avirulence and wide bands indicating virulence.
Virulence of A. tosichella on wheat genotypes containing Cmc2, Cmc3, or Cmc4 A. tosichella resistance genes, the Wsm2 Wheat Streak Mosaic Virus resistance gene plus Cmc4, the OK05312 Cmc4 mite-resistant control and the susceptible Jagger control at 14 dpi by A. tosichella populations collected from 25 counties in six U.S. Great Plains wheat producing states. Color bands represent A. tosichella numbers from each population on plants of each genotype. Narrow bands indicate avirulence, wide bands indicate virulence
Results
Plant leaf folding responses
Mites within each of the 25 populations caused variable responses in phenotypic leaf folding to mite-resistant and susceptible control plants. Feeding by mites from 15 of the 17 locations resulted in 0–20% leaf folding on OK05312 resistant control plants and 80–100% leaf folding on susceptible Jagger control plants (Table 1). Exceptions to this pattern occurred in mites from Dickinson, Finney and Geary co. Kansas, which caused 40–60% leaf folding on the susceptible control; and in mites from Pettis and Pike co. Missouri which caused 0–40% leaf folding on the resistant control and 60–80% folding on the susceptible control.
Plants containing Cmc4 and Wsm2 sustained significantly less leaf folding than susceptible Jagger control plants when infested with mites from Barton, Ellis, Ellsworth, Greeley, and Saline co. Kansas (Table 1). However, there were no statistical differences between resistant and susceptible controls and any test genotype fed on by mites from Dickinson, Finney, and Geary co. Kansas. Plants containing Cmc2 demonstrated significantly less leaf folding than susceptible Jagger control plants when infested with mites from Greeley co., and plants containing Cmc3 demonstrated a similar response when infested with mites from Barton and Greeley co.
In Missouri, the Cooper co. population caused significantly less folding to plants containing Cmc2, Cmc3, or Cmc4 + Wsmv2 than susceptible control plants (Table 1). Feeding by populations in Cape Girardeau and Stoddard co. also caused significantly less folding in Cmc4 + Wsmv2 plants than in susceptible control plants (Table 1). The Barton co. population caused significantly less folding in Cmc3 plants, but not to plants containing Cmc2 or Cmc4 +Wsmv2. There were no significant differences in leaf folding between resistant- and susceptible control plants or plants containing any of the resistance genes infested with mite populations from Pettis and Pike counties, Missouri.
Leaf folding by all Nebraska and South Dakota mite populations was significantly less on plants containing Cmc4 and Wsm2 than on susceptible plants and folding was no different than that on resistant plants (Table 1). Leaf folding caused by mites in Hughes co. South Dakota was significantly less on plants containing Cmc2 and Cmc3 than the susceptible plants; and folding caused by mites in Tripp co. South Dakota was significantly less on plants containing Cmc3 than on susceptible plants (Table 1). Mite populations in North Dakota and Texas caused significantly less folding to plants containing Cmc4 + Wsmv2 than the susceptible (Table 1), and both North Dakota populations caused significantly less folding to plants containing Cmc3.
Mite population counts
The mean mite populations on Cmc4 resistant- and susceptible control plants differed significantly when infested with mites from 20 of the 25 mite populations (Table 3, Supplemental Table 2). The five locations where there were no differences were Dickinson and Greeley co. Kansas; Cape Girardeau and Cooper co. Missouri; and Ward co. North Dakota. However, populations of mites from Cape Girardeau, Cooper, and Greeley co. on susceptible control plants were 3- to 10-fold higher than on Cmc4 resistant control plants (Table 2). Populations of mites from Dickinson co. Kansas did not differ on any plant genotype and were relatively reduced (16.6–68.6 mites/plant) (Table 2). The numbers of mites from Cape Girardeau and Cooper co. Missouri were significantly less on plants containing Cmc3 than susceptible controls, and populations of mites from Cape Girardeau were significantly lower on plants containing Cmc3 or Cmc4 + Wsm2 than susceptible controls (Table 2). Mites from Ward co. North Dakota exhibited a unique response (no differed significantly between Cmc4 resistant- and susceptible control). Finally, mite populations on plants containing Cmc4, Cmc4 + Wsm2 and the susceptible control did not differ, and all were significantly less than those on plants containing Cmc2 or Cmc3 (Table 2).
The gene Cmc2 was the least resistant of the Cmc genes assessed, with mites from only 6 of 25 locations (24%) exhibiting populations on Cmc2 plants significantly lower than those on susceptible control plants. Resistance in Cmc3 plants was comparatively broader. Mites from 14 of the 25 locations (56%) exhibited populations on Cmc3 plants significantly lower than those on susceptible control plants (Table 2). However, Cmc3 resistance was overcome by populations at two locations (Finney co. Kansas, Ward co. North Dakota), where mite populations were significantly higher on Cmc3 plants than on susceptible control plants. Plants containing the Cmc4 or the Cmc4 + Wsm2 combination displayed the broadest level of mite resistance. Counts of mites from 20 of 25 locations (80%) on Cmc4 plants or on Cmc4 + Wsm2 plants were significantly lower than those on susceptible control plants. The study found 17 of these 20 locations were identical (Table 2). The responses of mites from all populations to Cmc4 showed no indication of wide-scale mite virulence to Cmc4 (Table 3, Fig. 2). However, mite counts from Greeley and Dickinson co. in Kansas, and Cape Girardeau and Cooper co. in Missouri were no different on resistant- and susceptible controls, suggesting the potential existence of virulence to Cmc4. Nevertheless, the overall pattern of mite virulence was very similar to the trends in mite population abundance shown in Table 2.
Virus infection
WSMV was the most prevalent virus detected in mites in all 25 mite populations and was present in 75% of the populations (Supplemental Table 4). The HPWMoV and TriMV were much less prevalent, present in only 16% and 13% of the populations, respectively. The WSMV was also present at significantly greater levels than HPWMoV or TriMV in mites collected from 18 of the 25 locations (Table 4, Supplemental Table 4). There few multiple infections transmitted by mites from the 25 different locations sampled, although WSMV/HPWMoV coinfections and WSMV/TriMV coinfections were detected in occurred in 11% and 12% of all sample locations, respectively (Supplemental Table 4). The WSMV/HPWMoV coinfection occurred only in South Dakota (53%), North Dakota (40%), and at a much lower incidence of 3% in Missouri (Supplemental Table 4). The WSMV/TriMV coinfection occurred at 50% of the Missouri sample sites, 20% of Nebraska sample sites, at only 3% of the sites sampled in Kansas. No HPWMoV/TriMV coinfections or triple WSMV/HPWMoV/TriMV coinfections were detected in plants infested with mites from any sample site in our experiments (Supplemental Table 4). In addition, WSMV infection and HPWMoV infection in mites collected from Bottineau co. North Dakota occurred at significantly greater levels than TriMV infection. Finally, WSMV and TriMV infections in mites from Barton and Pike co. Missouri occurred at significantly greater levels than infection by HPWMoV (Table 4, Supplemental Table 2).
ELISA ratios of the three viruses differed significantly in plants infested by mites from 22 of these locations (Table 4). At the remaining three locations there no significant differences in the ELISA ratios of plants infected by mites from Geary co. Kansas, Pettis co. Missouri, or Saunders co. Nebraska. The WSMV ELISA ratio was significantly greater than the HPWMoV or TriMV ELISA ratios in plants infected by mites in six of the seven counties in Kansas and all other counties in Missouri and Nebraska. The HPWMoV ELISA ratio was significantly greater than WSMV or TriMV ratios in plants infected by mites in Saline co. and Ward co. North Dakota. Finally, WSMV and HPWMoV ELISA were ratios significantly greater than TriMV ratios in plants infected by mites from Lake and Tripp co. South Dakota. HPWMoV was only present in plants infected by mites in North and South Dakota and TriMV was only present in plants infected by mites in Barton and Pike co. Missouri, and Furnas co. Nebraska (Supplemental Table 4).
Discussion
The presence of a wheat curl mite biotype with the ability to overcome antibiosis resistance controlled by the Cmc3 gene in wheat cultivar TAM107 was documented > 20 years ago (Harvey et al. 1999). This plasticity in mite physiology has since served as a reminder that a thorough knowledge of mite virulence patterns is an essential element of effective mite management in the U.S. Great Plains. Our results demonstrated that Cmc4 resistant plants exhibit only 0–20% leaf folding at the regional level, suggesting that Cmc4 remains effective in the Great Plains. Similarly, plants of the KSU2R-2 breeding line containing Cmc4 and Wsm2 also showed significantly less leaf folding than susceptible Jagger plants in response to all mite populations except those in three counties in Kansas and three counties in Missouri (Table 1). These responses are similar to those determined by Chuang et al. (2017) with KSU2R-2. Mite population reductions resulting from feeding on breeding line KSU2R-2 paralleled leaf folding scores on KSU2R-2 plants, where mite populations were significantly lower than those on susceptible plants in 20 of 25 populations sampled (Table 2).
Interestingly, plants containing Cmc2 from PI52452 and Cmc3 from TAM107 displayed significantly less leaf folding to mite populations from Greeley co. Kansas, Cooper co. Missouri, and Hughes co. South Dakota, suggesting that these genes remain effective sources of mite resistance in some locations (Table 1). Plants containing Cmc3 also displayed significantly less leaf folding in response to feeding by mites from Tripp co. South Dakota, and Bottineau and Ward co. North Dakota (Table 1). Taken together, these results demonstrate that Cmc2- and Cmc3—based resistance also remains effective against several mite populations in the U.S. Great Plains.
The lack of widespread occurrence of additional A. tosichella virulence since 1996 may also be related to the genetic content of wheat varieties planted in the Great Plains. Since the occurrence of virulence to Cmc3, several varieties with A. tosichella- or WSMV resistance have been developed and currently constitute from 30 to 60% of the varieties grown in Colorado, Kansas, Oklahoma and Texas (Hunger et al. 2018; Neely et al. 2018; Pachl et al. 2017). During this same ~ 20 year period, new varieties containing Cmc4 have also been released and are now grown in Kansas and Oklahoma (OK052312), and in Montana (Yellowstone) (Hofer et al. 2011; Carver et al. 2016).
The WSMV was predominant among all mite populations, occurring at a rate of 75%, compared to HPWMoV or TriMV, which were transmitted by mites from only 16% and 13% of all locations, respectively. Viral infection rates in wheat plants collected in the Spring of 2008 in Kansas, Nebraska, and South Dakota were also previously reported by Burrows et al. (2009) and by Byamukama et al. (2013) for plants collected in the Spring of 2011 and 2012. However, their results were based on ELISA data from field-collected plants and derived from much greater numbers of samples (~ 750 in 2008, ~ 5000 in 2011 and 2012), compared to the 125 plant samples in our viral transmission experiments. Nevertheless, WSMV infection rates in Kansas and Nebraska in 2008, 2011 and 2012 were similar to our results, ranging from 50 to 75% in Kansas and from 39 to 50% in Nebraska. In contrast, our experiments detected a 93% rate of WSMV infection in plants transmitted by mites from South Dakota, compared to only 28% plant infection in 2008 (Burrows et al. 2009) and 18% plant infection in 2011 and 2012 (Byamukama et al. 2013).
Burrows et al. (2009) and Byamukama et al. (2013) also determined lower overall rates of HPWMoV or TriMV infection than WSMV, but there was considerable variation between our results for these viruses and those of Burrows et al. (2009) and Byamukama et al. (2013). Our experiments detected 60% HPWMoV infection in plants infested with mites from South Dakota, whereas Burrows et al. (2009) and Byamukama et al. (2013) detected < 1% infection. In contrast, our results and those of Byamukama et al. (2013) detected < 10% HPWMoV infection in Kansas, compared to 38% HPWMoV infection detected by Burrows et al. (2009).
Our results also differed from those of Burrows et al. (2009) and Byamukama et al. (2013) in detection of double- and triple coinfections of the three viruses. Mites collected from Kansas and Nebraska in our experiments transmitted no WSMV/HPWMoV coinfections or a very low incidence of WSMV/TriMV coinfections (Supplemental Table 4). In contrast, Burrows et al. (2009) and Byamukama et al. (2013) detected these coinfections at rates ranging from 5 to 15% WSMV/HPWMoV and 18 to 30% WSMV/TriMV. Our experiments also detected a significant rate of 53% WSMV/HPWMoV coinfection in South Dakota in 2016, compared to > 10% coinfection in 2008, 2011 or 2012 (Burrows et al. 2009, Byamukama et al. 2013).
Single infection rates for WSMV (93%) or HPWMoV (60%) that we observed from mites collected in South Dakota in 2016 (Supplemental Table 4) were much higher than those observed in 2008 by Burrows et al. (2009) (28% WSMV, 7% HPWMoV) or those observed in 2011 and 2012 by Byamukama et al. (2013) (18% WSMV, < 1% HPWMoV). Thus, it is tempting to suggest this major increase in the presence of both WSMV and HPWMoV in South Dakota over a recent 12-year period to be related to global temperature increase. This conclusion is premature; however, until improved geo-temporal-spatial models are available that incorporate information on virus infection rates, vector presence, vector virus competency, wheat variety, and vegetative ground cover at the local and regional levels.
Our results provide no evidence to support the contention that virulence is correlated to a greater incidence of WSMV, HPWMoV, or TriMV. The populations from Copper co. Missouri and Ward co. North Dakota were both virulent to Cmc2, Cmc4, and Cmc4 + Wsm2 (Table 3), yet each population vectored a completely different virus. Mites from Copper co. transmitted a significantly greater level of WSMV than those from Ward co. and mites from and Ward co. transmitted a significantly greater level of HPWMoV than those from Copper co. (Table 4).
Wheat curl mite populations in our study were taken in each of the same eight counties in Kansas sampled by Harvey et al. (1999) in 1997. Comparisons of previous and current responses of mites at these locations indicate that virulence to Cmc2 increased or remained the same in six of eight counties and diminished from virulence to intermediate (not different from resistant or susceptible control plant response) in two counties (Table 3). Virulence to Cmc3 has increased only in mites from Finney co., remained the same in Barton, Ellis, and Saline cos., and diminished from virulent to intermediate in mites from all other Kansas counties. Taken together, our results demonstrate wheat curl mite virulence to Cmc3 in Kansas has diminished since 1997 and increased slightly to Cmc2.
In general, our results suggest that wheat varieties containing Cmc4 or Cmc4 + Wsm2 are strong sources of mite resistance with excellent potential for use in U.S. Great Plains wheat breeding programs. However, the intermediate levels of avirulence to Cmc4 in four mite populations sampled in Kansas, Missouri, and Nebraska reinforce the need for continued mite virulence monitoring in areas of chronically high wheat curl mite populations. Results of our experiments represent the first survey of the U.S. Great Plans wheat production area since 1999 for A. tosichella virulence and the first to assess the incidence mite-transmitted infection of wheat by Wheat streak mosaic virus; High Plains wheat mosaic virus; and Triticum Mosaic Virus since 2008.
References
Andrews JE, Slykhuis JT (1956) Reaction of winter wheat varieties and Triticum × Agropyron hybrids when inoculated with wheat streak mosaic virus by the mite vector Aceria tulipae Keifer. Plant Dis Rep 40:513–516
Atkinson TG, Grant MN (1967) An evaluation of streak mosaic losses in winter wheat. Phytopathol 57:188–192
Burrows M, Franc G, Rush C, Blunt T, Ito D, Kinzer K, Ziems A (2009) Occurrence of viruses in wheat in the Great Plains region, 2008. Plant Health Prog. https://doi.org/10.1094/PHP-2009-0706-01-RS
Byamukama E, Seifers DL, Hein GL, De Wolf E, Tisserat NA, Langham MAC, Osborne LE, Timmerman A, Wegulo SN (2013) Occurrence and distribution of Triticum mosaic virus in the central Great Plains. Plant Dis 97:21–29
Byamukama E, Tatineni S, Hein GL, McMechan J, Wegulo SN (2016) Incidence of Wheat streak mosaic virus, Triticum mosaic virus and Wheat mosaic virus in wheat curl mites recovered from maturing winter wheat spikes. Plant Dis 100:318–323
Carver BF, Smith CM, Chuang W-P, Hunger RM, Edwards JT, Yan L, Bowden RL (2016) Registration of OK05312, a high-yielding hard winter wheat donor of for wheat curl mite resistance. J Plant Regist 10:75–79
Christian ML, Willis WG (1993) Survival of wheat streak mosaic virus in grass hosts in Kansas for wheat harvest to fall wheat emergence. Plant Dis 77:239–242
Chuang W-P, Aguirre-Rojas LM, Khalaf LK, Zhang G, Fritz AK, Whitfield AE, Smith CM (2017) Wheat genotypes with combined resistance to wheat curl mite, wheat streak mosaic virus, wheat mosaic virus, and triticum mosaic virus. J Econ Entomol 110:711–718
Conner RL, Thomas JB, Whelan EDP (1991) Comparison of mite resistance for control of wheat streak mosaic. Crop Sci 31:315–318
Connin RV (1956) The host range of the wheat curl mite, vector of wheat streak-mosaic. J Econ Entomol 49:1–4
DeWolf ED, Bockus WW, Whitworth J (2014) Wheat variety disease and insect ratings 2014. Agricultural Experiment Station and Cooperative Extension Service, Kansas State University, Manhattan
Fahim M, Mechanicos A, Ayala-Navarrete L, Haber S, Larkin PJ (2012) Resistance to Wheat streak mosaic virus—a survey of resources and development of molecular markers. J Plant Pathol 61:425–440
Fisher RA (1954) Statistical methods for research worker. Oliver and Boyd, Edinburgh
French R, Stenger DC (2003) Evolution of Wheat streak mosaic virus: dynamics of population growth within plants may explain limited variation. Annu Rev Phytopathol 41:199–214
Graybosch RA, Peterson CJ, Baenziger PS, Baltensperger DD, Nelson LA, Jin Y, Martin TJ (2009) Registration of ‘Mace’ hard red winter wheat. J Plant Regist 3:51–56
Harvey TL, Livers RL (1975) Resistance to wheat curl mite, Aceria tulipae Keifer, in rye and wheat-rye addition lines. Environ Entomol 4:523–526
Harvey TL, Martin TJ (1980) Effects of wheat pubescence on infestations of wheat curl mite and incidence of wheat streak mosaic. J Econ Entomol 73:225–227
Harvey TL, Martin TJ (1988) Sticky-tape method to measure cultivar effect on wheat curl mite (Acari: Eriophyidae) populations in wheat spikes. J Econ Entomol 81:731–734
Harvey TL, Martin TJ, Seifers DL (1990) Wheat curl mite and wheat streak mosaic in moderate trichome density wheat cultivars. Crop Sci 30:534–536
Harvey TL, Martin TJ, Seifers DL (1994) Importance of plant resistance to insect and mite vectors in controlling virus diseases of plants: resistance to the wheat curl mite (Acari: Eriophyidae). J Agric Entomol 11:271–277
Harvey TL, Martin TJ, Seifers DL (1995) Survival of five wheat curl mite, Aceria tosichella Keifer (Acari: Eriophyidae), strains on mite resistant wheat. Exp Appl Acarol 19:459–463
Harvey TL, Martin TJ, Seifers DL, Sloderbeck PE (1997) Change in virulence of wheat curl mite detected on TAM 107 wheat. Crop Sci 37:624–625
Harvey TL, Seifers DL, Martin TJ, Brown-Guedira G, Gill BS (1999) Survival of wheat curl mites on different sources of resistance in wheat. Crop Sci 39:1887–1889
Harvey TL, Seifers DL, Martin TJ, Michaud JP (2005) Effect of resistance to wheat streak mosaic virus on transmission efficiency of wheat curl mites. J Agric Urban Entomol 22:1–6
Hofer P, Berg JE, Huang L, Graf RJ, Bruckner PL (2011) Registration of ‘Yellowstone’ winter wheat backcross—derived lines incorporating leaf rust and wheat curl mite resistance. J Plant Regist 5:422–425
Hunger B, Marburger D, Carver B, Royer T (2018) Wheat variety comparison June 2018. Oklahoma Cooperative Extension Service. PSS-2142
Kobayashi T (2016) Evolving ideas about genetics underlying insect virulence to plant resistance in rice-brown planthopper interactions. J Insect Physiol 84:32–39
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marral MA (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645. https://doi.org/10.1101/gr.092759.109
Liu W, Seifers DL, Qi L, Friebe B, Gill B (2011) A compensating wheat—Robertsonian translocation conferring resistance to and mosaic virus. Crop Sci 51:2382–2390
Lu H, Price J, Devkota, Rush C, Rudd J (2011) A dominant gene for resistance to in winter wheat line CO960293-2. Crop Sci 51:5–12
Mahmood T, Hein GL, Jensen SG (1998) Mixed infection of hard red winter wheat with high plains virus and wheat streak mosaic virus from wheat curl mites in Nebraska. Plant Dis 82:311–315
Malik R (2001) Molecular genetic characterization of wheat curl mite, Aceria tosichella Keifer (Acari: Eriophyidae), and wheat genes conferring wheat curl mite resistance. PhD Dissertation, Kansas State University
Malik R, Brown-Guedira GL, Smith CM, Harvey TL, Gill BS (2003) Genetic mapping of wheat curl mite resistance genes and in common wheat. Crop Sci 43:644–650
Martin TJ, Harvey TL, Livers RW (1976) Resistance to wheat streak mosaic virus and its vector, Aceria tulipae. Phytopathol 66:346–349
Martin TJ, Harvey TL, Bender CG, Seifers DL, Hatchett JH (1983) Wheat curl mite resistant wheat germplasm. Crop Sci 23:809
Martin TJ, Harvey TL, Bender CG, Seifers DL, Hatchett JH (1984) Control of wheat streak mosaic virus with vector resistance in wheat. Phytopathology 74:963–964
McMechan AJ, Hein GL (2016) Planting date and variety selection for management of viruses transmitted by the wheat curl mite (Acari: Eriophyidae). J Econ Entomol 109:70–77
Milliken G, Johnson D (2004) Analysis of messy data: design of experiments, vol 1, 2nd edn. Chapman and Hall, CRC Press, Boca Raton
Morgan G, Patrick C, Steddom K, Rush C (2005) Wheat streak mosaic virus and High Plains virus. Texas Cooperative Extension Publ. E-337
Murugan M, Cardona PS, Duraimurugan P, Whitfield AE, Schneweis D, Smith CM (2011) Wheat curl mite resistance: interactions of mite feeding with wheat streak mosaic virus infection. J Econ Entomol 104:1406–1414
Neely C, Gerrish B, Hathcoat D, Drake D, Trostle C, Bell J, Kimura E, McGinty J, Ramirez J, Livingston C, Sirmon P, Ibrahim A, Rudd J, Sutton R, Devkota R, Baker J, Baker S, Opeña G, Simoneaux B, Jones CC, Braley A, Niece B. 2018 (2018) Texas wheat uniform variety trials. Texas A&M AgriLife Extension Service Publication SCS-2018-08. https://www.varietytesting.tamu.edu/wheat
O’Neal ME, Varenhorst AD, Kaiser MC (2018) Rapid evolution to host plant resistance by an invasive herbivore: soybean aphid (Aphis glycines) virulence in North America to aphid resistant cultivars. Curr Opin Insect Sci 26:1–7
Orlob GB (1966) Feeding and transmission characteristics of Aceria tulipae Keifer as vector of wheat streak mosaic virus. Phytopathol Z 55:218–238
Pachl C, Ott R, Meyer B (2017) Colorado winter wheat varieties 2018 crop. United States Department of Agriculture, National Agricultural Statistics Service. http://wheat.agsci.colostate.edu/wp-content/uploads/sites/85/2018/02/2018-Colorado-Survey.pdf
Rotenberg D, Bockus WW, Whitfield AE, Hervey K, Baker KD, Ou Z, Appel JA (2016) Occurrence of viruses and associated grain yields of paired symptomatic and nonsymptomatic tillers in Kansas winter wheat fields. Phytopathology 106:202–210
SAS (2008) The SAS system for windows. Release version 9.2. SAS institute, Cary, NC
Schlegel R, Kynast R (1987) Confirmation of a 1A/1R wheat-rye chromosome translocation in the wheat variety ‘Amigo’. Plant Breed 98:57–60
Seifers DL, Harvey TL, Martin TJ, Jensen SG (1997) Identification of the wheat curl mite as the vector of the High Plains virus of corn and wheat. Plant Dis 81:1161–1166
Seifers DL, Martin TJ, Harvey TL, Fellers JP, Stack JP, Ryba-White M, Yamchuk A (2008) Triticum mosaic virus: a new virus isolated from wheat in Kansas. Plant Dis 92:808–817
Seifers DL, Martin TJ, Harvey TL, Haber S, Krokhin O, Spicer V, Standing KG (2009a) Identification of variants of the High Plains virus infecting wheat in Kansas. Plant Dis 93:1265–1274
Seifers DL, Martin TJ, Harvey TL, Fellers JP, Michaud JP (2009b) Identification of the wheat curl mite as the vector of Triticum mosaic virus. Plant Dis 93:25–29
Shapiro SS, Francia R (1972) An approximate analysis of variance test for normality. J Am Stat Assoc 67:215–216
Sim T IV, Willis WG, Eversmeyer MG (1988) Kansas plant disease survey. Plant Dis 72:832–836
Skoracka A, Kuczyński L, De Mendonça RS, Dabert M, Szydło W, Knihinicki D, Navia D (2012) Cryptic species within the wheat curl mite Aceria tosichella (Keifer) (Acari: Eriophyoidea), revealed by mitochondrial, nuclear and morphometric data. Invertebr Syst 26:417–433
Slykhuis JT (1955) Aceria tulipae Keifer (Acarina: Eriophyidae) in relation to the spread of wheat streak mosaic. Phytopathology 45:116–128
Smith CM (1999) Plant resistance to insects. In: Rechcigl J, Rechcigl N (eds) Biological and biotechnological control of insect pests. CRC Press, Boca Raton, pp 171–205
Smith CM (2005) Plant resistance to arthropods: molecular and conventional approaches. Springer, Berlin
Smith CM, Clement SL (2012) Molecular bases of plant resistance to arthropods. Annu Rev Entomol 57:309–328
Thomas JB, Conner RL (1986) Resistance to colonization by the wheat curl mite in Aegilops squarrosa and its inheritance after transfer to common wheat. Crop Sci 26:527–530
Thomas JB, Conner RL, Graf RJ (2012) Radiant hard red winter wheat. Can J Plant Sci 92:169–175
Townsend L, Johnson D (1996) Wheat streak mosaic virus and the wheat curl mite (ENTFACT-117). University of Kentucky, KY
Triebe B, Mukai Y, Dhaliwal H, Martin T, Gill B (1991) Identification of alien chromatin specifying resistance to wheat streak mosaic and greenbug in wheat germ plasm by C-banding and in situ hybridization. Theor Appl Genet 81:381–389
USDA NASS (2013) U.S. Department of Agriculture, National Agricultural Statistics Service
Velandia M, Rejesus RM, Jones DC, Price JA, Workneh F, Rush CM (2010) Economic impact of wheat streak mosaic virus in the Texas high plains. Crop Prot 29:699–703
Whelan EDP, Hart GE (1988) A spontaneous translocation that transfers wheat curl mite resistance from decaploid Agropyron elongatum to common wheat. Genome 30:89–292
Whelan EDP, Thomas JB (1989) Chromosomal location in common wheat of a gene (Cmcl) from Aegilops squarrosa that conditions resistance to colonization by the wheat curl mite. Genome 32:1033–1036
Acknowledgements
The authors gratefully acknowledge the USDA/NIFA North Central IPM Program, the Kansas Wheat Commission, the Kansas Association of Wheat Growers, University of Baghdad, and Kansas State University Research and Extension for supporting this research. Contribution No. 19-128-J from the Kansas Agricultural Experiment Station.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling Editor: Joe Louis.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Khalaf, L., Chuang, WP., Aguirre-Rojas, L.M. et al. Differences in Aceria tosichella population responses to wheat resistance genes and wheat virus transmission. Arthropod-Plant Interactions 13, 807–818 (2019). https://doi.org/10.1007/s11829-019-09717-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11829-019-09717-9