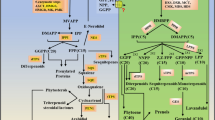
Abstract
S-nitrosylation is a well-known post-translational modification that modulates nitric oxide-dependent cell signaling. NADH-dependent S-nitrosoglutathione reductase (GSNOR) and NADPH-dependent thioredoxin reductase (NTR) enzymes are essential for nitric oxide/S-nitrosothiol (NO/SNO) homeostasis. GSNOR and NTR regulate denitrosylation by reducing S-nitrosoglutathione (GSNO) and thioredoxins, respectively. Genome-wide identification yielded 4 GSNOR and 12 NTR (4 each of NTRA, NTRB, and NTRC) genes in Brassica juncea. Syntenic relationship showed whole genome triplication (WGT) and tandem duplications. The phylogenetic analysis revealed clustering of BjGSNORs and BjNTRs with Arabidopsis homologs suggesting high sequence similarity within groups. Subcellular localization prediction suggested BjGSNOR localizes not only to the cytosol, but also to the Golgi apparatus and endoplasmic reticulum. BjNTRA and BjNTRB were localized in the cytoplasm and mitochondria, respectively, whereas BjNTRC localized in the chloroplast and nucleus. Several cis-acting elements involved in light responsiveness and expression analysis suggested the regulation of denitrosylation by light. The analysis of the promoter region also showed various phytohormone-regulated elements, suggesting the involvement of these enzymes in plant growth. Furthermore, GSNOR and NTR activities were higher in early growth stages. Differential spatial distributions of both the enzymes were observed with higher activity in hypocotyl in comparison with roots and cotyledons of the seedling. In flower, the highest activities were observed in carpel and least in stamens. Collectively, these findings provide an understanding of the structure, localization, and evolution of multiple copies of denitrosylases BjGSNOR and BjNTR, along with their possible roles in plant development.








Similar content being viewed by others
Data availability
The data sets supporting the results of this article are included within the article and its additional file.
References
Abat JK, Mattoo AK, Deswal R (2008) S-nitrosylated proteins of a medicinal CAM plant Kalanchoe pinnata–ribulose-1, 5-bisphosphate carboxylase/oxygenase activity targeted for inhibition. FEBS J 275(11):2862–2872. https://doi.org/10.1111/j.1742-4658.2008.06425.x
Airaki M, Sánchez-Moreno L, Leterrier M, Barroso JB, Palma JM, Corpas FJ (2011) Detection and quantification of S-nitrosoglutathione (GSNO) in pepper (Capsicum annuum L.) plant organs by LC-ES/MS. Plant Cell Physiol 52(11):2006–2015. https://doi.org/10.1093/pcp/pcr133
Airaki M, Leterrier M, Valderrama R, Chaki M, Begara-Morales JC, Barroso JB, del Río LA, Palma J, Corpas FJ (2015) Spatial and temporal regulation of the metabolism of reactive oxygen and nitrogen species during the early development of pepper (Capsicum annuum) seedlings. Ann Bot 116:679–693. https://doi.org/10.1093/aob/mcv023
Albertos P, Romero-Puertas MC, Tatematsu K, Mateos I, Sánchez-Vicente I, Nambara E, Lorenzo O (2015) S-nitrosylation triggers ABI5 degradation to promote seed germination and seedling growth. Nat Commun 6(1):1–10. https://doi.org/10.1038/ncomms9669
Alkhalfioui F, Renard M, Montrichard F (2007) Unique properties of NADP-thioredoxin reductase C in legumes. J Exp Bot 58(5):969–978. https://doi.org/10.1093/jxb/erl248
Arc E, Galland M, Godin B, Cueff G, Rajjou L (2013) Nitric oxide implication in the control of seed dormancy and germination. Front Plant Sci 4:346. https://doi.org/10.3389/fpls.2013.00346
Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2:28–36
Barroso JB, Valderrama R, Corpas FJ (2013) Immunolocalization of S-nitrosoglutathione, S-nitrosoglutathione reductase and tyrosine nitration in pea leaf organelles. Acta Physiol Plant 35:2635–2640. https://doi.org/10.1007/s11738-013-1291-0
Benhar M, Forrester MT, Hess DT, Stamler JS (2008) Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science 320(5879):1050–1054. https://doi.org/10.1126/science.1158265
Benhar M, Thompson JW, Moseley MA, Stamler JS (2010) Identification of S-nitrosylated targets of thioredoxin using a quantitative proteomic approach. Biochem 49(32):6963–6969. https://doi.org/10.1021/bi100619k
Berger H, De Mia M, Morisse S, Marchand CH, Lemaire SD, Wobbe L, Kruse O (2016) A light switch based on protein S-nitrosylation fine-tunes photosynthetic light harvesting in chlamydomonas. Plant Physiol 171:821–832. https://doi.org/10.1104/pp.15.0187
Blum M, Chang HY, Chuguransky S, Grego T, Kandasaamy S, Mitchell A, Finn RD (2021) The InterPro protein families and domains database: 20 years on. Nucleic Acids Res 49(D1):D344–D354. https://doi.org/10.1093/nar/gkaa977
Chatterji A, Sengupta R (2020) Cellular S-denitrosylases: potential role and interplay of thioredoxin, TRP14, and glutaredoxin systems in thiol-dependent protein denitrosylation. Int J Biochem Cell Biol 131:105904. https://doi.org/10.1016/j.biocel.2020.105904
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R (2020) TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13(8):1194–1202. https://doi.org/10.1016/j.molp.2020.06.009
Chen H, Wang T, He X, Cai X, Lin R, Liang J, Wang X (2022) BRAD V3.0: an upgraded Brassicaceae database. Nucleic Acids Res 50(D1):D1432–D1441. https://doi.org/10.1093/nar/gkab1057
Ciacka K, Krasuska U, Otulak-Kozieł K, Gniazdowska A (2019) Dormancy removal by cold stratification increases glutathione and S-nitrosoglutathione content in apple seeds. Plant Physiol Biochem 138:112–120. https://doi.org/10.1016/j.plaphy.2019.02.026
Correa-Aragunde N, Cejudo FJ, Lamattina L (2015) Nitric oxide is required for the auxin-induced activation of NADPH-dependent thioredoxin reductase and protein denitrosylation during root growth responses in Arabidopsis. Ann Bot 116(4):695–702. https://doi.org/10.1093/aob/mcv116
Dı́az M, Achkor H, Titarenko E, Martı́nez MC (2003) The gene encoding glutathione-dependent formaldehyde dehydrogenase/GSNO reductase is responsive to wounding, jasmonic acid and salicylic acid. FEBS Lett 543(1–3):136–139. https://doi.org/10.1016/S0014-5793(03)00426-5
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinfo 29(1):15–21. https://doi.org/10.1093/bioinformatics/bts635
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32(5):1792–1797. https://doi.org/10.1093/nar/gkh340
Espunya MC, Díaz M, Moreno-Romero J, Martínez MC (2006) Modification of intracellular levels of glutathione-dependent formaldehyde dehydrogenase alters glutathione homeostasis and root development. Plant Cell Environ 29(5):1002–1011. https://doi.org/10.1111/j.1365-3040.2006.01497.x
Feechan A, Kwon E, Yun BW, Wang Y, Pallas JA, Loake GJ (2005) A central role for S-nitrosothiols in plant disease resistance. Proc Natl Acad Sci USA 102(22):8054–8059. https://doi.org/10.1073/pnas.0501456102
Feng J, Chen L, Zuo J (2019) Protein S-nitrosylation in plants: current progresses and challenges. J Integr Plant Biol 61(12):1206–1223. https://doi.org/10.1111/jipb.12780
Frungillo L, De Oliveira JFP, Saviani EE, Oliveira HC, Martínez MC, Salgado I (2013) Modulation of mitochondrial activity by S-nitrosoglutathione reductase in Arabidopsis thaliana transgenic cell lines. Biochim Biophys Acta 1827(3):239–247. https://doi.org/10.1016/j.bbabio.2012.11.011
Gasteiger E, Hoogland C, Gattiker A, Wilkins MR, Appel RD, Bairoch A (2005) Protein identification and analysis tools on the ExPASy server. The proteomics protocols handbook. Springer, Berlin, pp 571–607. https://doi.org/10.1385/1-59259-890-0:571
Gupta KJ, Mur LAJ, Wany A, Kumari A, Fernie AR, Ratcliffe RG (2020) The role of nitrite and nitric oxide under low oxygen conditions in plants. New Phytol 225:1143–1151. https://doi.org/10.1111/nph.15969
Gupta KJ, Kaladhar VC, Fitzpatrick TB, Fernie AR, Møller IM, Loake GJ (2022) Nitric oxide regulation of plant metabolism. Mol Plant 15(2):228–242. https://doi.org/10.1016/j.molp.2021.12.012
Holmgren A, Bjornstedt M (1995) Thioredoxin and thioredoxin reductase. Methods in enzymology (252). Academic Press, Cambridge, pp 199–208. https://doi.org/10.1016/0076-6879(95)52023-6
Holzmeister C, Fröhlich A, Sarioglu H, Bauer N, Durner J, Lindermayr C (2011) Proteomic analysis of defense response of wildtype Arabidopsis thaliana and plants with impaired NO-homeostasis. Proteomics 11(9):1664–1683. https://doi.org/10.1002/pmic.201000652
Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinfo 31(8):1296–1297. https://doi.org/10.1093/bioinformatics/btu817
Hurali DT, Bhurta R, Tyagi S, Sathee L, Sandeep AB, Singh D, Mallick N, Vinod JSK (2022) Analysis of NIA and GSNOR family genes and nitric oxide homeostasis in response to wheat-leaf rust interaction. Sci Rep 12:803. https://doi.org/10.1038/s41598-021-04696-5
Hussain A, Yun B, Kim JH, Gupta KJ, Hyung N, Loake GJ (2019) Novel and conserved functions of S-nitrosoglutathione reductase in tomato. J Exp Bot 70(18):4877–4886. https://doi.org/10.1093/jxb/erz234
Jahnová J, Luhová L, Petřivalský M (2019) S-nitrosoglutathione reductase—the master regulator of protein S-nitrosation in plant NO signaling. Plants 8(2):48. https://doi.org/10.3390/plants8020048
Jedelská T, Luhová L, Petřivalský M (2020) Thioredoxins: emerging players in the regulation of protein S-nitrosation in plants. Plants 9(11):1426. https://doi.org/10.3390/plants9111426
Kalinina E, Novichkova M (2021) Glutathione in protein redox modulation through S-glutathionylation and S-nitrosylation. Molecules 26(2):435. https://doi.org/10.3390/molecules26020435
Kang Z, Qin T, Zhao Z (2019) Thioredoxins and thioredoxin reductase in chloroplasts: a review. Gene 706:32–42. https://doi.org/10.1016/j.gene.2019.04.041
Kneeshaw S, Gelineau S, Tada Y, Loake GJ, Spoel SH (2014) Selective protein denitrosylation activity of Thioredoxin-h5 modulates plant Immunity. Mol Cell 56(1):153–162. https://doi.org/10.1016/j.molcel.2014.08.003
Kolbert ZS, Barroso JB, Brouquisse R, Corpas FJ, Gupta KJ, Lindermayr C, Hancock JT (2019) A forty year journey: the generation and roles of NO in plants. Nitric Oxide 93:53–70. https://doi.org/10.1016/j.niox.2019.09.006
Kolbert Z, Lindermayr C, Loake GJ (2021) The role of nitric oxide in plant biology: current insights and future perspectives. J Exp Bot 72(3):777–780. https://doi.org/10.1093/jxb/erab013
Kubienová L, Kopečný D, Tylichová M, Briozzo P, Skopalová J, Šebela M, Petřivalský M (2013) Structural and functional characterization of a plant S-nitrosoglutathione reductase from Solanum lycopersicum. Biochimie 95(4):889–902. https://doi.org/10.1016/j.biochi.2012.12.009
Kubienová L, Tichá T, Jahnová J, Luhová L, Mieslerová B, Petřivalský M (2014) Effect of abiotic stress stimuli on S-nitrosoglutathione reductase in plants. Planta 239(1):139–146. https://doi.org/10.1007/s00425-013-1970-5
Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105–132. https://doi.org/10.1016/0022-2836(82)90515-0
Laha SD, Dutta S, Schäffner AR, Das M (2020) Gene duplication and stress genomics in Brassicas: current understanding and future prospects. J Plant Physiol 255:153293. https://doi.org/10.1016/j.jplph.2020.153293
Laloi C, Rayapuram N, Chartier Y, Grienenberger JM, Bonnard G, Meyer Y (2001) Identification and characterization of a mitochondrial thioredoxin system in plants. Proc Natl Acad Sci 98(24):14144–14149. https://doi.org/10.1073/pnas.241340898
Lawrence SR, Gaitens M, Guan Q, Dufresne C, Chen S (2020) S-nitroso-proteome revealed in stomatal guard cell response to flg22. Int J Mol Sci 5:1688. https://doi.org/10.3390/ijms21051688
Lee U, Wie C, Fernandez BO, Feelisch M, Vierling E (2008) Modulation of nitrosative stress by S-nitrosoglutathione reductase is critical for thermotolerance and plant growth in Arabidopsis. Plant Cell 20(3):786–802. https://doi.org/10.1105/tpc.107.052647
Lepisto A, Kangasjarvi S, Luomala EM, Brader G, Sipari N, Keranen M, Rintamaki E (2009) Chloroplast NADPH-thioredoxin reductase interacts with photoperiodic development in Arabidopsis. Plant Physiol 149(3):1261–1276. https://doi.org/10.1104/pp.108.133777
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30(1):325–327. https://doi.org/10.1093/nar/30.1.325
Lindermayr C (2018) Crosstalk between reactive oxygen species and nitric oxide in plants: key role of S-nitrosoglutathione reductase. Free Radical Bio Med 122:110–115. https://doi.org/10.1016/j.freeradbiomed.2017.11.027
Liu L, Hausladen A, Zeng M, Que L, Heitman J, Stamler JS (2001) A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature 410(6827):490–494. https://doi.org/10.1038/35068596
Lopes-Oliveira PJ, Oliveira HC, Kolbert Z, Freschi L (2021) The light and dark sides of nitric oxide: multifaceted roles of nitric oxide in plant responses to light. J Exp Bot 72(3):885–903. https://doi.org/10.1093/jxb/eraa504
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. https://doi.org/10.1186/s13059-014-0550-8
Ma Z, Marsolais F, Bykova NV, Igamberdiev AU (2016) Nitric oxide and reactive oxygen species mediate metabolic changes in barley seed embryo during germination. Front Plant Sci 7:138. https://doi.org/10.3389/fpls.2016.00138
Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Bryant SH (2015) CDD: NCBI’s conserved domain database. Nucleic Acids Res 43(D1):D222–D226. https://doi.org/10.1093/nar/gku1221
Matamoros MA, Cutrona MC, Wienkoop S, Begara-Morales JC, Sandal N, Orera I, Becana M (2020) Altered plant and nodule development and protein S-nitrosylation in Lotus japonicus mutants deficient in S-nitrosoglutathione reductases. Plant Cell Physiol 61(1):105–117. https://doi.org/10.1093/pcp/pcz182
Mata-Pérez C, Spoel SH (2019) Thioredoxin-mediated redox signalling in plant immunity. Plant Sci 279:27–33. https://doi.org/10.1016/j.plantsci.2018.05.001
Mathur S, Paritosh K, Tandon R, Pental D, Pradhan AK (2022) Comparative analysis of seed transcriptome and coexpression analysis reveal candidate genes for enhancing seed size/weight in Brassica juncea. Front Genet 13:814486. https://doi.org/10.3389/fgene.2022.814486
Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer EL, Bateman A (2021) Pfam: the protein families database in 2021. Nucleic Acids Res 49(D1):D412–D419. https://doi.org/10.1093/nar/gkaa913
Nakai K (1999) PSORT: a program for detecting the sorting signals of proteins and predicting their subcellular localization. Trends Biochem Sci 24(1):34–35. https://doi.org/10.1016/S0968-0004(98)01336-X
Ojeda V, Pérez-Ruiz JM, González M, Nájera VA, Sahrawy M, Serrato AJ, Cejudo FJ (2017) NADPH thioredoxin reductase C and thioredoxins act concertedly in seedling development. Plant Physiol 174(3):1436–1448. https://doi.org/10.1104/pp.17.00481
Paritosh K, Yadava SK, Singh P, Bhayana L, Mukhopadhyay A, Gupta V, Bisht NC, Zhang J, Kudrna DA, Copetti D, Wing RA, Reddy Lachagari VB, Pradhan AK, Pental D (2021) A chromosome-scale assembly of allotetraploid Brassica juncea (AABB) elucidates comparative architecture of the A and B genomes. Plant Biotechnol J 19(3):602–614. https://doi.org/10.1111/pbi.13492
Pérez-Ruiz JM, González M, Spínola MC, Sandalio LM, Cejudo FJ (2009) The quaternary structure of NADPH thioredoxin reductase C is redox-sensitive. Mol Plant 2(3):457–467. https://doi.org/10.1093/mp/ssp011
Prakash V, Singh VP, Tripathi DK, Sharma S, Corpas FJ (2019) Crosstalk between nitric oxide (NO) and abscisic acid (ABA) signalling molecules in higher plants. Environ Exp Bot 161:41–49. https://doi.org/10.1016/j.envexpbot.2018.10.033
Reichheld JP, Khafif M, Riondet C, Droux M, Bonnard G, Meyer Y (2007) Inactivation of thioredoxin reductases reveals a complex interplay between thioredoxin and glutathione pathways in Arabidopsis development. Plant Cell 19(6):1851–1865. https://doi.org/10.1105/tpc.107.050849
Reumann S, Babujee L, Ma C, Wienkoop S, Siemsen T, Antonicelli GE, Jahn O (2007) Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. Plant Cell 19(10):3170–3193. https://doi.org/10.1105/tpc.107.050989
Rizza S, Filomeni G (2017) Chronicles of a reductase: biochemistry, genetics and physio-pathological role of GSNOR. Free Radical Biol Med 110:19–30. https://doi.org/10.1016/j.freeradbiomed.2017.05.014
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–442. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Sakamoto A, Ueda M, Morikawa H (2002) Arabidopsis glutathione-dependent formaldehyde dehydrogenase is an S-nitrosoglutathione reductase. FEBS Lett 515(1–3):20–24. https://doi.org/10.1016/S0014-5793(02)02414-6
Savojardo C, Martelli PL, Fariselli P, Profiti G, Casadio R (2018) BUSCA: an integrative web server to predict subcellular localization of proteins. Nucleic Acids Res 46(W1):W459–W466. https://doi.org/10.1093/nar/gky320
Sehrawat A, Abat JK, Deswal R (2013) RuBisCO depletion improved proteome coverage of cold responsive S-nitrosylated targets in Brassica juncea. Front Plant Sci 4:342. https://doi.org/10.3389/fpls.2013.00342
Sehrawat A, Sougrakpam Y, Deswal R (2019) Cold modulated nuclear S-nitrosoproteome analysis indicates redox modulation of novel Brassicaceae specific, myrosinase and napin in Brassica juncea. Environ Exp Bot 161:312–333. https://doi.org/10.1016/j.envexpbot.2018.10.010
Serrato AJ, Pérez-Ruiz JM, Spínola MC, Cejudo FJ (2004) A novel NADPH thioredoxin reductase, localized in the chloroplast, which deficiency causes hypersensitivity to abiotic stress in Arabidopsis thaliana. J Biol Chem 279(42):43821–43827. https://doi.org/10.1074/jbc.M404696200
Shahpiri A, Svensson B, Finnie C (2008) The NADPH-dependent thioredoxin reductase/thioredoxin system in germinating barley seeds: gene expression, protein profiles, and interactions between isoforms of thioredoxin h and thioredoxin reductase. Plant Physiol 146(2):789. https://doi.org/10.1104/pp.107.113639
Shi YF, Wang DL, Wang C, Culler AH, Kreiser MA, Suresh J, Cohen JD, Pan J, Baker B, Liu JZ (2015) Loss of GSNOR1 function leads to compromised auxin signaling and polar auxin transport. Mol Plant 8:1350–1365. https://doi.org/10.1016/j.molp.2015.04.008
Signorelli S, Considine MJ (2018) Corrigendum: nitric oxide enables germination by a four-pronged attack on ABA-induced seed dormancy. Front Plant Sci 9:654. https://doi.org/10.3389/fpls.2018.00654
Sun H, Feng F, Liu J, Zhao Q (2018) Nitric oxide affects rice root growth by regulating auxin transport under nitrate supply. Front Plant Sci 9:659. https://doi.org/10.3389/fpls.2018.00659
Tada Y, Spoel SH, Pajerowska-Mukhtar K, Mou Z, Song J, Wang C, Zuo J, Dong X (2008) Plant immunity requires conformational changes of NPR1 via S-nitrosylation and thioredoxins. Science 321(5891):952–956. https://doi.org/10.1126/science.1156970
Tamura K, Stecher G, Kumar S (2021) MEGA 11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 38(7):3022–3027. https://doi.org/10.1093/molbev/msab120
Tichá T, Činčalová L, Kopečný D, Sedlářová M, Kopečná M, Luhová L, Petřivalský M (2017a) Characterization of S-nitrosoglutathione reductase from Brassica and Lactuca spp. and its modulation during plant development. Nitric Oxide 68:68–76. https://doi.org/10.1016/j.niox.2016.12.002
Tichá T, Lochman J, Činčalová L, Luhová L, Petřivalský M (2017b) Redox regulation of plant S-nitrosoglutathione reductase activity through post-translational modifications of cysteine residues. Biochem Biophys Res Commun 494(1–2):27–33. https://doi.org/10.1016/j.bbrc.2017.10.090
Wang P, Du Y, Hou YJ, Zhao Y, Hsu CC, Yuan F, Zhu JK (2015) Nitric oxide negatively regulates abscisic acid signaling in guard cells by S-nitrosylation of OST1. Proc Natl Acad Sci 112(2):613–618. https://doi.org/10.1073/pnas.1423481112
Wen D, Gong B, Sun S, Liu S, Wang X, Wei M, Yang F, Li Y, Shi Q (2016) Promoting roles of melatonin in adventitious root development of Solanum lycopersicum L. by regulating auxin and nitric oxide signaling. Fron Plant Sci. https://doi.org/10.3389/fpls.2016.00718
Wünsche H, Baldwin IT, Wu J (2011) S-Nitrosoglutathione reductase (GSNOR) mediates the biosynthesis of jasmonic acid and ethylene induced by feeding of the insect herbivore Manduca sexta and is important for jasmonate-elicited responses in Nicotiana attenuata. J Exp Bot 62(13):4605–4616. https://doi.org/10.1093/jxb/err171
Xu S, Guerra D, Lee U, Vierling E (2013) S-nitrosoglutathione reductases are low-copy number, cysteine-rich proteins in plants that control multiple developmental and defense responses in Arabidopsis. Front Plant Sci 4:430. https://doi.org/10.3389/fpls.2013.00430
Yokochi Y, Fukushi Y, Wakabayashi KI, Yoshida K, Hisabori T (2021) Oxidative regulation of chloroplast enzymes by thioredoxin and thioredoxin-like proteins in Arabidopsis thaliana. Proc Natl Acad Sci. https://doi.org/10.1073/pnas.2114952118
Yu M, Lamattina L, Spoel SH, Loake GJ (2014) Nitric oxide function in plant biology: a redox cue in deconvolution. New Phytol 202(4):1142–1156. https://doi.org/10.1111/nph.12739
Zuccarelli R, Coelho AC, Peres LE, Freschi L (2017) Shedding light on NO homeostasis: Light as a key regulator of glutathione and nitric oxide metabolisms during seedling deetiolation. Nitric Oxide 68:77–90. https://doi.org/10.1016/j.niox.2017.01.006
Acknowledgements
This work was partially supported by the FRP grant provided by Institute of Eminence, University of Delhi [IOE/2021/12/FRP] and SAP grant F.3-5/2018/DRS-II (SAP-II). PB availed senior research fellowship from UGC, India.
Funding
University of Delhi, IOE/2021/12/FRP, Renu Deswal, University Grant Commission, F.3-5/2018/DRS-II (SAP-II), Renu Deswal, NET 331105 (2014), Priyanka Babuta.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Babuta, P., Paritosh, K. & Deswal, R. Genome-wide identification and functional analysis of denitrosylases (S-nitrosoglutathione reductases and NADPH-dependent thioredoxin reductases) in Brassica juncea. Plant Biotechnol Rep 17, 541–559 (2023). https://doi.org/10.1007/s11816-023-00831-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11816-023-00831-y