Abstract
The initiation of flowering is a crucial event in the life cycle of plants. Flowering marks the transition of the plant from its vegetative to reproductive state. Flowers are shoot modifications derived from flower primordia, as a means of reproduction and securing seed production adopted by plants to transmit their genomic information across generations for the survival of the species. Floral transition is a consequence of the interplay between endogenous factors such as plant genetic structure, and exogenous factors such as photoperiod, temperature, and nutrients. The generic mechanism of flowering initiation is evolutionarily conserved across plant families. In addition, it is highly dependent on the genetic and physiological characteristics of individual species, in coordination with the surrounding environmental factors. Therefore, flowering control is extremely adaptable to seasonal changes, ensuring the reproductive success of the plant species. The genes involved in flowering control maintain a delicate balance orchestrated by different signalling mechanisms in response to the environmental cues. In this study, various mechanisms related to flowering are collectively demonstrated in terms of endogenous genetic cues responding to natural exogenous triggers and artificial inducers, including environmental factors and phytohormones, respectively.




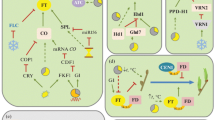
source: https://onlinelibrary.wiley.com/doi/full/10.1046/j.1365-313X.2003.01833.x (Moon et al. 2003)
Similar content being viewed by others
Abbreviations
- AP1,2:
-
APATELLA 1,2
- PI:
-
Pistillata
- AG:
-
Agamous
- TF1:
-
Terminal flower 1
- LFY:
-
Leafy gene
- CAL:
-
Cauliflower
- FUL:
-
Fruitful
- UFO:
-
Unusual floral organs
- EF1:
-
Elongation factor 1
- FT:
-
Flowering locus T
- CO:
-
Constans
- TSF1:
-
Twin sister of FT
- F-BOX1:
-
F-box transcription factor
- FKF1:
-
Flavin-binding Kelch repeat, F-box1
- LOV:
-
Light, oxygen, voltage
- GI:
-
Gigantea
- CDF1:
-
Cyclin DOF factor 1
- COP1:
-
Constitutive photomorphogenic 1
- PRC 1&2:
-
Polycomb repressive complexes 1,2
- NF-Y:
-
Nuclear factor Y
- SAM:
-
Shoot apical meristem
- FD:
-
Flowering locus D
- FDP:
-
FD paralog
- FLC:
-
Flowering locus D
- MADS:
-
Initial letter if MCM1 agamous, deficiens
- SOC1:
-
Suppressor of overexpression of CO1
- COI1:
-
Coronatine insensitive 1
- SPL15:
-
Squamosa promoter-binding-like protein 15
- FCA:
-
Flowering control locus A
- FY:
-
Flowering time control protein Y
- FLK:
-
Flowering locus KH domain
- LD:
-
Long day
- FLD:
-
Flowering locus D
- SD:
-
Short day
- ZTL:
-
Zeitlupe
- LKP2:
-
LOV Kelch protein 2
- CCA1:
-
Circadian clock associated 1
- LHY:
-
Late elongated hypocotyl
- TOC1:
-
Timing of CAB expression 1
- LUX:
-
Lux Arrythmo
- ELF4:
-
Early flowering 4
- EC:
-
Evening complex
- PIF:
-
Phytochrome interacting factor
- SVP:
-
Short vegetative phase, HOS1, high expression of osmotically responsive genes 1
- ABA:
-
Abscisic acid
- ABRE:
-
ABA-responsive element binding protein
- ABFs:
-
ABRE-binding factors
- FMT:
-
Friendly mitochondria
- TFL1:
-
Terminal flower1
- ABI3:
-
Abscisic acid insensitive 3
- GAI:
-
GA insensitive;
- RGA:
-
Repressor of GA1-3
- RGL1:
-
RGA-like 1
- NTL8:
-
Transmembrane motif 1- like 8
- BFT:
-
Brother of FT
- CRY1:
-
Cryptochrome 1
- JA:
-
Jasmonic acid
- ET:
-
Ethylene
- RGL123:
-
RGA-like 1,2,3
- SOC1:
-
MADS-box gene SOC1
- MYB33:
-
Myeloblastosis 33
- PIN1:
-
Peptidylproyl cis/trans isomerase NIMA interacting 1
- PAT:
-
Phosphinothricin acetyl transferase
- AIL:
-
Aintegumenta-like
- PLT:
-
Plethora
- ERF:
-
Ethylene-responsive factor
- PYR:
-
Pyrabactin resistance
- RCAR:
-
Regulatory components of ABA receptors
- PP2Cs:
-
Type 2 C protein phosphatases
- SnRK:
-
Snf1- related protein kinase
- bZIP:
-
Basic leucine zipper
- ABI:
-
Abscisic acid insensitive
- AAC:
-
1-Aminocyclopropane-1-carboxylic acid
- ACS:
-
AAC synthase
- EIN3:
-
Ethylene sensitive 3
- EIL:
-
EIN3- like
- ATR1:
-
Altered tryptophan regulator 1
- CTR1:
-
Coronatine insensitive 1
- EBF 12:
-
EIN3-binding f box protein 1
- SCFcoi1 E3:
-
SKP/Cullin1/F-box protein COI1 E3-ubiquitin-ligase complex
- JAZ:
-
JASMONATE ZIM domain
- bHLH-MYCs:
-
Basic helix–loop–helix-myelocytomatosis oncogenes
References
Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng J, Harberd NP (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science(new York, NY) 311:91
Achard P, Baghour M, Chapple A, Hedden P, Van Der Straeten D, Genschik P, Moritz T, Harberd NP (2007) The plant stress hormone ethylene controls floral transition via DELLA-dependent regulation of floral meristem-identity genes. Proc Natl Acad Sci USA 104:6484–6489
Alejandra Mandel M, Gustafson-Brown C, Savidge B, Yanofsky MF (1992) Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360:273–277
Álvarez-Aragón R, Haro R, Benito B, Rodríguez-Navarro A (2016) Salt intolerance in Arabidopsis: shoot and root sodium toxicity, and inhibition by sodium-plus-potassium overaccumulation. Planta 243:97–114
Amano T, Smithers RJ, Sparks TH, Sutherland WJ (2010) A 250-year index of first flowering dates and its response to temperature changes. Proc R Soc B 277:2451–2457
An H, Roussot C, Suárez-López P, Corbesier L, Vincent C, Piñeiro M, Hepworth S, Mouradov A, Justin S, Turnbull C, Coupland G (2004a) CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131:3615
An H, Roussot C, Suárez-López P, Corbesier L, Vincent C, Piñeiro M, Hepworth S, Mouradov A, Justin S, Turnbull C, Coupland G (2004b) Constans acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development (cambridge, England) 131:3615–3626
Balasubramanian S, Sureshkumar S, Lempe J, Weigel D (2006) Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet 2:e106
Bäurle I, Dean C (2008) Differential interactions of the autonomous pathway RRM proteins and chromatin regulators in the silencing of Arabidopsis targets. PLoS ONE 3:e2733
Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115:591–602
Chandler JW (2009) Local auxin production: a small contribution to a big field. BioEssays 31:60–70
Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR, Micol JL, Solano R (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448:666–671
Cho HJ, Kim JJ, Lee JH, Kim W, Jung JH, Park CM, Ahn JH (2012) Short vegetative phase (SVP) protein negatively regulates miR172 transcription via direct binding to the pri-miR172a promoter in Arabidopsis. FEBS Lett 586:2332–2337
Choi J, Hyun Y, Kang MJ, In Yun H, Yun JY, Lister C, Dean C, Amasino RM, Noh B, Noh YS, Choi Y (2009) Resetting and regulation of flowering locus C expression during Arabidopsis reproductive development. Plant J 57:918–931
Chuang T-H, Li K-H, Li P-F, Yang C-H (2018) Functional analysis of an APETALA1-like MADS box gene from Eustoma grandiflorum in regulating floral transition and formation. Plant Biotechnol Rep 12:115–125
Chung KS, Yoo SY, Yoo SJ, Lee JS, Ahn JH (2010) Brother of FT and TFL1 (BFT), a member of the FT/TFL1 family, shows distinct pattern of expression during the vegetative growth of Arabidopsis. Plant Signal Behav 5:1102–1104
Coen ES, Meyerowitz EM (1991) The war of the whorls: genetic interactions controlling flower development. Nature 353:31–37
Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, Coupland G (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science (new York, NY) 316:1030–1033
Covington MF, Harmer SL (2007) The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol 5:e222
Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61:651–679
de Folter S, Immink RG, Kieffer M, Parenicová L, Henz SR, Weigel D, Busscher M, Kooiker M, Colombo L, Kater MM, Davies B, Angenent GC (2005) Comprehensive interaction map of the Arabidopsis MADS Box transcription factors. Plant Cell 17:1424–1433
Deng W, Ying H, Helliwell CA, Taylor JM, Peacock WJ, Dennis ES (2011) Flowering locus C (FLC) regulates development pathways throughout the life cycle of Arabidopsis. Proc Natl Acad Sci USA 108:6680–6685
Dill A, Sun T-P (2001a) Synergistic derepression of Gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159:777
Dill A, Sun T-P (2001b) Synergistic derepression of Gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159:777
Esmon CA, Tinsley AG, Ljung K, Sandberg G, Hearne LB, Liscum E (2006) A gradient of auxin and auxin-dependent transcription precedes tropic growth responses. Proc Natl Acad Sci USA 103:236–241
Fang Y, Xiong L (2015) General mechanisms of drought response and their application in drought resistance improvement in plants. Cell Mol Life Sci 72:673–689
Fernández V, Takahashi Y, Le Gourrierec J, Coupland G (2016) Photoperiodic and thermosensory pathways interact through Constans to promote flowering at high temperature under short days. Plant J 86:426–440
Ferrandiz C, Gu Q, Martienssen R, Yanofsky MF (2000a) Redundant regulation of meristem identity and plant architecture by Fruitfull, Apetala1 and Cauliflower. Development 127:725
Ferrandiz C, Gu Q, Martienssen R, Yanofsky MF (2000b) Redundant regulation of meristem identity and plant architecture by Fruitfull, Apetala1 and Cauliflower. Development 127:725–734
Galvão VC, Collani S, Horrer D, Schmid M (2015) Gibberellic acid signaling is required for ambient temperature-mediated induction of flowering in Arabidopsis thaliana. Plant J 84:949–962
Gälweiler L, Guan C, Müller A, Wisman E, Mendgen K, Yephremov A, Palme K (1998) Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science (new York, NY) 282:2226
Griffiths J, Murase K, Rieu I, Zentella R, Zhang Z-L, Powers SJ, Gong F, Phillips AL, Hedden P, Sun T-P, Thomas SG (2006) Genetic characterization and functional analysis of the GID1 Gibberellin receptors in Arabidopsis. Plant Cell 18:3399
Gu X, Le C, Wang Y, Li Z, Jiang D, Wang Y, He Y (2013) Arabidopsis FLC clade members form flowering-repressor complexes coordinating responses to endogenous and environmental cues. Nat Commun 4:1947
Guo H, Ecker JR (2003) Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115:667–677
Gusmaroli G, Tonelli C, Mantovani R (2001) Regulation of the CCAAT-Binding NF-Y subunits in Arabidopsis thaliana. Gene 264:173–185
He Y, Michaels SD, Amasino RM (2003) Regulation of flowering time by histone acetylation in Arabidopsis. Science (new York, NY) 302:1751–1754
Henderson IR, Liu F, Drea S, Simpson GG, Dean C (2005) An allelic series reveals essential roles for FY in plant development in addition to flowering-time control. Development (cambridge, England) 132:3597–3607
Heo JB, Sung S (2011) Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science (new York, NY) 331:76
Hornyik C, Terzi LC, Simpson GG (2010) The spen family protein FPA controls alternative cleavage and polyadenylation of RNA. Dev Cell 18:203–213
Huang H, Nusinow DA (2016) Into the evening: complex interactions in the Arabidopsis circadian clock. Trends Genet 32:674–686
Huang W, Pérez-García P, Pokhilko A, Millar AJ, Antoshechkin I, Riechmann JL, Mas P (2012) Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science (new York, NY) 336:75–79
Hwang K, Susila H, Nasim Z, Jung J-Y, Ahn JH (2019) Arabidopsis ABF3 and ABF4 transcription factors act with the NF-YC complex to regulate SOC1 expression and mediate drought-accelerated flowering. Mol Plant 12:489–505
Jagadish SV, Bahuguna RN, Djanaguiraman M, Gamuyao R, Prasad PV, Craufurd PQ (2016) Implications of high temperature and elevated CO2 on flowering time in plants. Front Plant Sci 7:913
Jiang C, Gao X, Liao L, Harberd NP, Fu X (2007) Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the gibberellin-DELLA signaling pathway in Arabidopsis. Plant Physiol 145:1460–1470
Jiang D, Wang Y, Wang Y, He Y (2008) Repression of Flowering Locus C and Flowering Locus T by the Arabidopsis polycomb repressive complex 2 components. PLoS ONE 3:e3404
Jung JH, Seo PJ, Park CM (2012) The E3 ubiquitin ligase HOS1 regulates Arabidopsis flowering by mediating CONSTANS degradation under cold stress. J Biol Chem 287:43277–43287
Kant S, Peng M, Rothstein SJ (2011) Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in Arabidopsis. PLoS Genet 7:e1002021
Khanale V, Bhattacharya A, Satpute R, Char B (2021) Brief bioinformatics identification of cotton bZIP transcription factors family from Gossypium hirsutum, Gossypium arboreum and Gossypium raimondii. Plant Biotechnol Rep 15:493–511
Kim SG, Park CM (2007) Membrane-mediated salt stress signaling in flowering time control. Plant Signal Behav 2:517–518
Kim S, Choi K, Park C, Hwang H-J, Lee I (2006) Suppressor of FRIGIDA4, encoding a C2H2-Type zinc finger protein, represses flowering by transcriptional activation of Arabidopsis Flowering Locus C. Plant Cell 18:2985–2998
Kim J-S, Seo S-G, Jun B-K, Lee Y, Jeon SB, Choe J, Kim J-B, Kim ST, Kim S-H (2011) An IbEF1 from sweet potato promotes flowering in transgenic tobacco. Genes Genom 33:335–341
Kim JJ, Lee JH, Kim W, Jung HS, Huijser P, Ahn JH (2012) The microRNA156-squamosa promoter binding protein-LIKE3 module regulates ambient temperature-responsive flowering via Flowering Locus T in Arabidopsis. Plant Physiol 159:461–478
Kleine-Vehn J, Dhonukshe P, Sauer M, Brewer PB, Wiśniewska J, Paciorek T, Benková E, Friml J (2008) ARF GEF-dependent transcytosis and polar delivery of PIN auxin carriers in Arabidopsis. Curr Biol 18:526–531
Koornneef M, Alonso-Blanco C, Blankestijn-de Vries H, Hanhart CJ, Peeters AJ (1998) Genetic interactions among late-flowering mutants of Arabidopsis. Genetics 148:885–892
Krizek B (2009) Aintegumenta and Aintegumenta-Like6 act redundantly to regulate Arabidopsis floral growth and patterning. Plant Physiol 150:1916
Kumar SV, Wigge PA (2010) H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140:136–147
Kumar SV, Lucyshyn D, Jaeger KE, Alós E, Alvey E, Harberd NP, Wigge PA (2012) Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484:242–245
Kumimoto RW, Adam L, Hymus GJ, Repetti PP, Reuber TL, Marion CM, Hempel FD, Ratcliffe OJ (2008) The nuclear factor Y subunits NF-YB2 and NF-YB3 play additive roles in the promotion of flowering by inductive long-day photoperiods in Arabidopsis. Planta 228:709–723
la Rosa NM-d, Sotillo B, Miskolczi P, Gibbs DJ, Vicente J, Carbonero P, Oñate-Sánchez L, Holdsworth MJ, Bhalerao R, Alabadí D, Blázquez MA (2014) Large-scale identification of Gibberellin-related transcription factors defines group VII Ethylene Response Factors as functional DELLA partners. Plant Physiol 166:1022–1032
Langridge J (1957) Effect of day-length and Gibberellic acid on the flowering of Arabidopsis. Nature 180:36–37
Lee I, Aukerman MJ, Gore SL, Lohman KN, Michaels SD, Weaver LM, John MC, Feldmann KA, Amasino RM (1994) Isolation of LUMINIDEPENDENS: a gene involved in the control of flowering time in Arabidopsis. Plant Cell 6:75–83
Lee H, Yoo SJ, Lee JH, Kim W, Yoo SK, Fitzgerald H, Carrington JC, Ahn JH (2010) Genetic framework for flowering-time regulation by ambient temperature-responsive miRNAs in Arabidopsis. Nucleic Acids Res 38:3081–3093
Lee JH, Ryu H-S, Chung KS, Posé D, Kim S, Schmid M, Ahn JH (2013) Regulation of temperature-responsive flowering by MADS-Box transcription factor repressors. Science 342:628–632
Li K, Wang Y, Han C, Zhang W, Jia H, Li X (2007) GA signaling and CO/FT regulatory module mediate salt-induced late flowering in Arabidopsis thaliana. Plant Growth Regul 53:195–206
Liu H, Stone SL (2010a) Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell 22:2630–2641
Liu H, Stone SL (2010b) Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell 22:2630–2641
Liu F, Quesada V, Crevillén P, Bäurle I, Swiezewski S, Dean C (2007) The Arabidopsis RNA-binding protein FCA requires a lysine-specific demethylase 1 homolog to downregulate FLC. Mol Cell 28:398–407
Liu T, Li Y, Ren J, Qian Y, Yang X, Duan W, Hou X (2013) Nitrate or NaCl regulates floral induction in Arabidopsis thaliana. Biologia 68:215–222
Lutz U, Posé D, Pfeifer M, Gundlach H, Hagmann J, Wang C, Weigel D, Mayer KF, Schmid M, Schwechheimer C (2015) Modulation of ambient temperature-dependent flowering in Arabidopsis thaliana by natural variation of Flowering Locus M. PLoS Genet 11:e1005588
Macknight R, Bancroft I, Page T, Lister C, Schmidt R, Love K, Westphal L, Murphy G, Sherson S, Cobbett C, Dean C (1997) FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell 89:737–745
Mandel MA, Yanofsky MF (1995) A gene triggering flower formation in Arabidopsis. Nature 377:522–524
Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF (1992) Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360:273–277
Mockler TC, Yu X, Shalitin D, Parikh D, Michael TP, Liou J, Huang J, Smith Z, Alonso JM, Ecker JR, Chory J, Lin C (2004) Regulation of flowering time in Arabidopsis by K homology domain proteins. Proc Natl Acad Sci USA 101:12759–12764
Moon J, Suh S-S, Lee H, Choi K-R, Hong CB, Paek N-C, Kim S-G, Lee I (2003) The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J 35:613–623
Nole-Wilson S, Tranby TL, Krizek BA (2005) AINTEGUMENTA-like (AIL) genes are expressed in young tissues and may specify meristematic or division-competent states. Plant Mol Biol 57:613–628
Okada K, Ueda J, Komaki MK, Bell CJ, Shimura Y (1991) Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 3:677–684
Okamuro JK, Caster B, Villarroel R, Van Montagu M, Jofuku KD (1997) The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc Natl Acad Sci USA 94:7076–7081
Paltiel J, Amin R, Gover A, Ori N, Samach A (2006) Novel roles for GIGANTEA revealed under environmental conditions that modify its expression in Arabidopsis and Medicago truncatula. Planta 224:1255–1268
Parcy F, Nilsson O, Busch MA, Lee I, Weigel D (1998) A genetic framework for floral patterning. Nature 395:561–566
Pauwels L, Goossens A (2011) The JAZ proteins: a crucial interface in the Jasmonate signaling cascade. Plant Cell 23:3089
Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF (2000) B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405:200–203
Posé D, Verhage L, Ott F, Yant L, Mathieu J, Angenent GC, Immink RG, Schmid M (2013) Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature 503:414–417
Riboni M, Galbiati M, Tonelli C, Conti L (2013) GIGANTEA enables drought escape response via abscisic acid-dependent activation of the Florigens and Suppressor of Overexpression of Constans. Plant Physiol 162:1706
Riboni M, Robustelli Test A, Galbiati M, Tonelli C, Conti L (2016) ABA-dependent control of GIGANTEA signalling enables drought escape via up-regulation of Flowering Locus T in Arabidopsis thaliana. J Exp Bot 67:6309–6322
Rodríguez-Cazorla E, Ripoll JJ, Andújar A, Bailey LJ, Martínez-Laborda A, Yanofsky MF, Vera A (2015) K-homology nuclear ribonucleoproteins regulate floral organ identity and determinacy in Arabidopsis. PLoS Genet 11:e1004983
Ryu JY, Park C-M, Seo PJ (2011) The floral repressor Brother of FT and TFL1 (BFT) modulates flowering initiation under high salinity in Arabidopsis. Mol Cells 32:295
Salisbury FB, Marinos NG (1985) The ecological role of plant growth substances. In: Pharis RP, Reid DM (eds) Hormonal regulation of development III: role of environmental factors. Springer, Berlin Heidelberg, pp 707–766
Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carré IA, Coupland G (1998) The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93:1219–1229
Schomburg FM, Patton DA, Meinke DW, Amasino RM (2001) FPA, a gene involved in floral induction in Arabidopsis, encodes a protein containing RNA-recognition motifs. Plant Cell 13:1427–1436
Shang Y, Yan L, Liu ZQ, Cao Z, Mei C, Xin Q, Wu FQ, Wang XF, Du SY, Jiang T, Zhang XF, Zhao R, Sun HL, Liu R, Yu YT, Zhang DP (2010) The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 22:1909–1935
Shannon S, Meeks-Wagner DR (1991) A mutation in the Arabidopsis TFL1 gene affects inflorescence meristem development. Plant Cell 3:877–892
Shin JM, Seo S-G, Kim J-S, Jang H, Shin M-R, Lee C, Kim S-H (2015) Ectopic expression of NtEF1 and NtEF2 promotes flowering and alters floral organ identity in Nicotiana tabacum. Plant Biotechnol Rep 9:11–26
Shu K, Chen Q, Wu Y, Liu R, Zhang H, Wang S, Tang S, Yang W, Xie Q (2016) Abscisic acid-insensitive 4 negatively regulates flowering through directly promoting Arabidopsis Flowering Locus C transcription. J Exp Bot 67:195–205
Simpson GG, Dijkwel PP, Quesada V, Henderson I, Dean C (2003) FY is an RNA 3’ end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell 113:777–787
Siriwardana NS, Lamb RS (2012) The poetry of reproduction: the role of LEAFY in Arabidopsis thaliana flower formation. Int J Dev Biol 56:207–221
Song YH, Smith RW, To BJ, Millar AJ, Imaizumi T (2012) FKF1 conveys timing information for Constans stabilization in photoperiodic flowering. Science (new York, NY) 336:1045–1049
Song J, Irwin J, Dean C (2013) Remembering the prolonged cold of winter. Curr Biol 23:R807–R811
Suárez-López P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G (2001) Constans mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410:1116–1120
Talbert PB, Henikoff S (2014) Environmental responses mediated by histone variants. Trends Cell Biol 24:642–650
Tejos R, Rodriguez-Furlán C, Adamowski M, Sauer M, Norambuena L, Friml J (2018) PATELLINS are regulators of auxin-mediated PIN1 relocation and plant development in Arabidopsis thaliana. J Cell Sci 131(2)
Tiwari SB, Shen Y, Chang HC, Hou Y, Harris A, Ma SF, McPartland M, Hymus GJ, Adam L, Marion C, Belachew A, Repetti PP, Reuber TL, Ratcliffe OJ (2010) The flowering time regulator Constans is recruited to the Flowering Locus T promoter via a unique cis-element. New Phytol 187:57–66
Vishwakarma K, Upadhyay N, Kumar N, Yadav G, Singh J, Mishra RK, Kumar V, Verma R, Upadhyay RG, Pandey M, Sharma S (2017) Abscisic acid signaling and abiotic stress tolerance in plants: a review on current knowledge and future prospects. Front Plant Sci 8
Wada KC, Takeno K (2010) Stress-induced flowering. Plant Signal Behav 5:944–947
Wagner D, Sablowski RWM, Meyerowitz EM (1999) Transcriptional activation of APETALA1 by Leafy. Science (new York, NY) 285:582
Wang ZY, Tobin EM (1998) Constitutive expression of the Circadian clock associated 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93:1207–1217
Wang KLC, Li H, Ecker JR (2002) Ethylene biosynthesis and signaling networks. Plant Cell 14:S131
Wang Y, Li L, Ye T, Lu Y, Chen X, Wu Y (2013) The inhibitory effect of ABA on floral transition is mediated by ABI5 in Arabidopsis. J Exp Bot 64:675–684
Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM (1992) LEAFY controls floral meristem identity in Arabidopsis. Cell 69:843–859
Wigge PA (2013) Ambient temperature signalling in plants. Curr Opin Plant Biol 16:661–666
Wilson RN, Heckman JW, Somerville CR (1992) Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol 100:403–408
Woodward G, Perkins DM, Brown LE (2010) Climate change and freshwater ecosystems: impacts across multiple levels of organization. Philos Transact R Soc B 365:2093–2106
Wu G, Poethig RS (2006) Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development (cambridge, England) 133:3539–3547
Xi W, Liu C, Hou X, Yu H (2010) MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. Plant Cell 22:1733–1748
Yoshida T, Mogami J, Yamaguchi-Shinozaki K (2015) Omics approaches toward defining the comprehensive abscisic acid signaling network in plants. Plant Cell Physiol 56:1043–1052
Yu H, Xu Y, Tan EL, Kumar PP (2002) AGAMOUS-LIKE 24, a dosage-dependent mediator of the flowering signals. Proc Natl Acad Sci 99:16336–16341
Yuan S, Zhang Z-W, Zheng C, Zhao Z-Y, Wang Y, Feng L-Y, Niu G, Wang C-Q, Wang J-H, Feng H, Xu F, Bao F, Hu Y, Cao Y, Ma L, Wang H, Kong D-D, Xiao W, Lin H-H, He Y (2016) Arabidopsis cryptochrome 1 functions in nitrogen regulation of flowering. Proc Natl Acad Sci 113:7661–7666
Zhai Q, Zhang X, Wu F, Feng H, Deng L, Xu L, Zhang M, Wang Q (2015) Transcriptional mechanism of Jasmonate receptor COI1-mediated delay of flowering time in Arabidopsis. Plant Cell 27:2814–2828
Zhang X, Garreton V, Chua NH (2005) The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev 19:1532–1543
Zhang S, Jin Y, Hao H, Liang S, Ma X, Luan W (2020) Characterization and identification of OsFTL8 gene in rice. Plant Biotechnol Rep 14:683–694
Acknowledgements
This research was funded by the New Breeding Technologies Development Program (No. PJ01485802), Rural Development Administration, Republic of Korea. The permission to use Fig. 4 in this manuscript has been granted (Moon et al. 2003).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kaur, C., Lim, Y.P. & Lee, GJ. Co-ordinated responses to endogenous and environmental triggers allow a well-timed floral transition in plants. Plant Biotechnol Rep 16, 145–159 (2022). https://doi.org/10.1007/s11816-021-00731-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11816-021-00731-z