Abstract
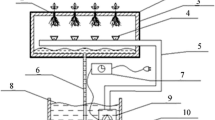
Plants respond to combined stresses with different phenotypic and transcriptional changes from those in single-stress situations. Increasing temperature and carbon dioxide (CO2) concentration are key climate change factors that could affect plant growth and development, but the molecular mechanisms regulating responses to these factors are still poorly understood. To broadly survey genes with altered expression during combined CO2 and heat stresses, RNA samples were prepared from needles of Korean fir subjected to high CO2 and heat. RNA-sequencing analyses revealed that the expression of a large number of transcripts was altered under high temperature. Intriguingly, the transcriptomic results showed fewer gene expression changes under a combination of both high CO2 and temperature compared with heat treatment alone. Gene ontology analysis revealed the differentially expressed transcripts were mainly associated with metabolic process, binding and cell terms. The expression profiles of transcripts involved in metabolic pathways and cellular responses were identified using MapMan analysis, which revealed that CO2 and heat stresses induced transcript expression related to light reactions, biotic and abiotic stress responses, and development. Additionally, transcription factor genes known to be important for abiotic stress responses, such as ERF, bHLH, and NAC, were identified. The reliability of the observed expression patterns was confirmed by quantitative RT-PCR. The genetic knowledge acquired here will be useful for future studies of the molecular adaptation of this tree species to simultaneous environmental stresses.





Similar content being viewed by others
References
Abebe A, Pathak H, Singh SD, Bhatia A, Harit RC, Kumar V (2016) Growth, yield and quality of maize with elevated atmospheric carbon dioxide and temperature in north-west India. Agric Ecosyst Environ 218:66–72
Ainsworth EA, Rogers A, Vodkin LO, Walter A, Schurr U (2006) The effects of elevated CO2 concentration on soybean gene expression. An analysis of growing and mature leaves. Plant Physiol 142:135–147
Arrhenius S (1896) On the influence of carbonic acid in the air upon the temperature of the ground. Phil Mag 41:237–276
Audic S, Claverie JM (1997) The significance of digital gene expression profiles. Genome Res. 7:986–995
Balasubramanian S, Sureshkumar S, Lempe JD (2006) Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet 2:e106
De Souza AP, Gaspar M, Da Silva EA, Ulian EC, Waclawovsky AJ, Nishiyama MY Jr, Dos Santos RV, Teixeira MM, Souza GM, Buckeridge MS (2008) Elevated CO2 increases photosynthesis, biomass and productivity, and modifies gene expression in sugarcane. Plant Cell Environ. 31:1116–1127
Dietz KJ, Vogel MO, Viehhauser A (2010) AP2/EREBP transcription factors are part of gene regulatory networks and integrate metabolic, hormonal and environmental signals in stress acclimation and retrograde signalling. Protoplasma 245:3–14
Feder ME, Mitchell-Olds T (2003) Evolutionary and ecological functional genomics. Nat Rev Genet 4:649–655
Fernandez-Caballero C, Rosales R, Romero I, Escribano MI, Merodio C, Sanchez-Ballesta MT (2012) Unraveling the roles of CBF1, CBF4 and dehydrin 1 genes in the response of table grapes to high CO2 levels and low temperature. J Plant Physiol 169:744–748
Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, Fox SE, Wong WK, Mockler TC (2010) A genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res 20:45–58
Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talón M, Dopazo J, Conesa A (2008) High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36:3420–3435
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652
Gray WM, Östin A, Sandberg G, Romano CP, Estelle M (1998) High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci 95:7197–7202
He F, Liu Q, Zheng L, Cui Y, Shen Z, Zheng L (2015) RNA-Seq analysis of rice roots reveals the involvement of post-transcriptional regulation in response to cadmium stress. Front Plant Sci 6:1136
Hsieh EJ, Cheng MC, Lin TP (2013) Functional characterization of an abiotic stress-inducible transcription factor AtERF53 in Arabidopsis thaliana. Plant Mol Biol 82:223–237
Hwang JE, Kim YJ, Shin MH, Hyun HJ, Bohnert HJ, Park HC (2018) A comprehensive analysis of the Korean Fir (Abies koreana) genes expressed under heat stress using transcriptome analysis. Sci reports 8:10233
Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D'Angelo C, Bornberg-Bauer E, Kudla J, Harter K (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50:347–363
Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper JF (2002) Transcriptome changes in Arabidopsis in response to salt, osmotic and cold stress. Plant Physiol 130:2129–2141
Langridge P, Paltridge N, Fincher G (2006) Functional genomics of abiotic stress tolerance in cereals. Brief Funct Genomic Proteomic 4:343–354
Lee TB (1982) Endemic plants and their distribution in Korea. J Natl Acad Sci 21:169–218
Lee SW, Yang BH, Han SD, Song JH, Lee JJ (2008) Genetic variation in natural populations of Abies nephrolepis Max. in South Korea. Ann For Sci 65:302
Li P, Ainsworth EA, Leakey AD, Ulanov A, Lozovaya V, Ort DR, Bohnert HJ (2008) Arabidopsis transcript and metabolite profiles: ecotype-specific responses to open-air elevated [CO2]. Plant Cell Environ 31:1673–1687
Lindemose S, O’Shea C, Jensen MK, Skriver K (2013) Structure, function and networks of transcription factors involved in abiotic stress responses. Int J Mol Sci 14:5842–5878
Mittler R (2006) Abiotic stress, the field environment and stress combination. Trends Plant Sci 11:15–19
Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2012) NAC transcription factors in plant abiotic stress responses. Biochim Biophys Acta 1819:97–103
Niu Y, Zhao T, Xu X, Li J (2017) Genome-wide identification and characterization of GRAS transcription factors in tomato (Solanum lycopersicum). PeerJ 5:e3955
Nuruzzaman M, Sharoni AM, Kikuchi S (2013) Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front Microbiol 4:248
Qiu Q, Ma T, Hu Q, Liu B, Wu Y, Zhou H, Wang Q, Wang J, Liu J (2011) Genome-scale transcriptome analysis of the desert poplar, Populus euphratica. Tree Physiol 31:452–461
Shahnejat-Bushehri S, MuellerRoeber B, Balazadeh S (2012) Arabidopsis NAC transcription factor JUNGBRUNNEN1 affects thermomemory-associated genes and enhances heat stress tolerance in primed and unprimed conditions. Plant Signal Behav 7:1518–1521
Song Y, Chen Q, Ci D, Shao X, Zhang D (2014a) Effects of high temperature on photosynthesis and related gene expression in poplar. BMC Plant Biol 14:111
Song Y, Ci D, Tian M, Zhang DQ (2014b) Comparison of the physiological effects and transcriptome responses of Populus simonii under different abiotic stresses. Plant Mol Biol 86:139–156
Springer CJ, Orozco RA, Kelly JK, Ward JK (2008) Elevated CO2 influences the expression of floral-initiation genes in Arabidopsis thaliana. New Phytol 178:63–67
Sreenivasulu N, Sopory SK, Kavi Kishor PB (2007) Deciphering the regulatory mechanisms of abiotic stress tolerance in plants by genomic approaches. Gene 388:1–13
Tallis MJ, Lin Y, Rogers A, Zhang J, Street NR, Miglietta F, Karnosky DF, De Angelis P, Calfapietra C, Taylor G (2010) The transcriptome of Populus in elevated CO reveals increased anthocyanin biosynthesis during delayed autumnal senescence. New Phytol. 186:415–428
Taub DR, Seemann JR, Coleman JS (2000) Growth in elevated CO2 protects photosynthesis against high-temperature damage. Plant Cell Environ 23:649–656
Taylor G, Ranasinghe S, Bosac C, Gardner SDL, Ferris R (1994) Elevated CO2 and plant growth: cellular mechanisms and responses of whole plants. J Exp Bot 45:1761–1774
Taylor G, Tricker PJ, Zhang FZ, Alston VJ, Miglietta F, Kuzminsky E (2003) Spatial and temporal effects of free-air CO2 enrichment (POPFACE) on leaf growth, cell expansion, and cell production in a closed canopy of poplar. Plant Physiol 131:177–185
Teng N, Wang J, Chen T, Wu X, Wang Y, Lin J (2006) Elevated CO2 induces physiological, biochemical and structural changes in leaves of Arabidopsis thaliana. New Phytol 172:92–103
Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37:914–939
Torres-Galea P, Hirtreiter B, Bolle C (2013) Two GRAS proteins, SCARECROW-LIKE21 and PHYTOCHROME A SIGNAL TRANSDUCTION1, function cooperatively in phytochrome A signal transduction. Plant Physiol 161:291–304
Wang H, Wang H, Shao H, Tang X (2016) Recent advances in utilizing transcription factors to improve plant abiotic stress tolerance by transgenic technology. Front Plant Sci 7:67
Watson R, Houghton J, Yihui D (2001) Climate change 2001: the scientific basis. Intergovernmental Panel on Climate Change, Geneva
Watson-Lazowski A, Lin Y, Miglietta F, Edwards RJ, Chapman MA, Taylor G (2016) Plant adaptation or acclimation to rising CO2? Insight from first multigenerational RNA-Seq transcriptome. Glob Chang Biol 22:3760–3773
Woo SY (2009) Forest decline of the world: a linkage with air pollution and global warming. Afr J Biotechnol 8:7409–7414
Xu H, Chen L, Song B, Fan X, Yuan X, Chen J (2016) De novo transcriptome sequencing of pakchoi (Brassica rapa L. chinensis) reveals the key genes related to the response of heat stress. Acta Physiol Plant 38:252
Yokotani N, Ichikawa T, Kondou Y, Matsui M, Hirochika H, Iwabuchi M, Oda K (2009) Tolerance to various environmental stresses conferred by the salt-responsive rice gene ONAC063 in transgenic Arabidopsis. Planta 229:1065–1075
Yu J, Yang Z, Jespersen D, Huang B (2014) Photosynthesis and protein metabolism associated with elevated CO2-mitigation of heat stress damages in tall fescue. Environ Exp Bot 99:75–85
Acknowledgements
This research was supported by the National Institute of Ecology (NIE-C-2019-15).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hwang, J.E., Kim, Y.J., Jeong, D.Y. et al. Transcriptome analysis of Korean fir (Abies koreana) in response to elevated carbon dioxide and high temperature. Plant Biotechnol Rep 13, 603–612 (2019). https://doi.org/10.1007/s11816-019-00553-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11816-019-00553-0