Abstract
Syringin, sinapyl alcohol 4-O-glucoside, is well known as a plant-derived bioactive monolignol glucoside. In Arabidopsis, recombinant chimeric protein UGT72E3/2 has been previously reported to lead to significantly higher syringin production than the parental enzymes UGT72E2 and UGT72E3. To enhance syringin content in Korean soybean (Glycine max L. ‘Kwangan’), we cloned the UGT72E3/2 gene under the control of the β-conglycinin or CaMV-35S promoter to generate β-UGT72E3/2 and 35S-UGT72E3/2 constructs, respectively, and then transformed them into soybean to obtain transgenic plants using the modified half-seed method. Real-time semi-quantitative PCR (RT-PCR) analysis showed that the UGT72E3/2 gene was expressed in the leaves of the β-UGT72E3/2 and 35S-UGT72E3/2 transgenic lines. HPLC analysis of the seeds and mature tissues of the T2 generation plants revealed that the β-UGT72E3/2 transgenic seeds accumulated 0.15 µmol/g DW of total syringin and 0.29 µmol/g DW of total coniferin, whereas coniferin and syringin were not detected in non-transgenic seeds. Moreover, coniferin and syringin also accumulated at high levels in non-seed tissues, particularly the leaves of β-UGT72E3/2 transgenic lines. In contrast, 35S-UGT72E3/2 lines showed no differences in the contents of coniferin and syringin between transgenic and non-transgenic soybean plants. Thus, the seed-specific β-conglycinin promoter might be an effective tool to apply to the nutritional enhancement of soybean crops through increased syringin production.




Similar content being viewed by others
References
Anterola AM, Lewis NG (2002) Trends in lignin modification: a comprehensive analysis of the effects of genetic manipulations/mutations on lignification and vascular integrity. Phytochemistry 61:221–294
Choi J, Shin KM, Park HJ, Jung HJ, Kim HJ, Lee YS, Rew JH, Lee KT (2004) Anti-inflammatory and antinociceptive effects of sinapyl alcohol and its glucoside syringin. Planta Med 11:1027–1032
Chu Y, Kwon T, Nam J (2014) Enzymatic and metabolic engineering for efficient production of syringin, sinapyl alcohol 4-O-glucoside, in Arabidopsis thaliana. Phytochemistry 102:55–63
Di R, Purcell V, Collins GB, Ghabrial SA (1996) Production of transgenic lines expressing the bean pod mottle virus coat protein precursor gene. Plant Cell Rep 15:746–750
Hinchee MAW, Connor-Ward DV, Newell CA, McDonnell RE, Sato SJ, Gasser CS, Fischhoff DA, Re DB, Fraley RT, Horsch RB (1988) Production of transgenic soybean plants using Agrobacterium-mediated DNA transfer. Nat Biotechnol 6:915–922
Jiang N, Jeon EH, Pak JH, Ha TJ, Baek IY, Jung WS, Lee JH, Kim DH, Choi HK, Cui Z, Chung YS (2010) Increase of isoflavones in soybean callus by Agrobacterium-mediated transformation. Plant Biotechnol Rep 4:253–260
John KMM, Natarajan S, Luthria DL (2016) Metabolite changes in nine different soybean varieties grown under field and greenhouse conditions. Food Chem 211:347–355
Karimi M, Inze´ D, Depicker A (2002) Gateway™ vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7:193–195
Kim NY, Pae HO, Ko YS, Yoo JC, Choi BM, Jun CD, Chung HT, Inagaki M, Higuchi R, Kim YC (1999) In vitro inducible nitric oxide synthesis inhibitory active constituents from Fraxinus rhynchophylla. Planta Med 65:656–658
Kim MJ, Kim JK, Kim HJ, Pak JH, Lee JH, Kim DH, Choi HK, Jung HW, Lee JD, Chung YS, Ha SH (2012) Genetic modification of the soybean to enhance the β-carotene content through seed-specific expression. PLoS One 7:e48287
Kim HJ, Kim MJ, Pak JH, Jung HW, Choi HK, Lee YH, Baek IY, Ko JM, Jeong SC, Pack IS, Ryu KH, Chung YS (2013) Characterization of SMV resistance of soybean produced by genetic transformation of SMV-CP gene in RNAi. Plant Biotechnol Rep 7:425–433
Kim HJ, Kim MJ, Pak JH, Im HH, Lee DH, Kim KH, Lee JH, Kim DH, Choi HK, Jung HW, Chung YS (2016) RNAi-mediated soybean mosaic virus (SMV) resistance of a Korean soybean cultivar. Plant Biotechnol Rep 10:257–267
Kim MJ, Kim HJ, Pak JH, Cho HS, Choi HK, Jung HW, Lee DH, Chung YS (2017) Overexpression of AtSZF2 from Arabidopsis showed enhanced tolerance to salt stress in soybean. Plant Breed Biotech 5:1–15
Krishnan SSC, Subramanian IP, Subramanian SP (2014) Isolation, characterization of syringin, phenylpropanoid glycoside from Musa paradisiaca tepal extract and evaluation of its antidiabetic effect in streptozotocin-induced diabetic rats. Biomed Prevent Nutr 4:105–111
Lanot A, Hodge D, Jackson RG, George GL, Elias L, Lim EK, Vaistij FE, Bowles DJ (2006) The glucosyltransferase UGT72E2 is responsible for monolignol 4-O-glucoside production in Arabidopsis thaliana. Plant J 48:286–295
Lanot A, Hodge D, Lim EK, Vaistij FE, Bowles DJ (2008) Redirection of flux through the phenylpropanoid pathway by increased glucosylation of soluble intermediates. Planta 228:609–616
Leinhos V, Savidge RA (1993) Isolation of protoplasts from developing xylem of Pinus banksiana and Pinus strobus. Can J For Res 23:343–348
Lim EK, Li Y, Parr A, Jackson R, Ashford DA, Bowles DJ (2001) Identification of glucosyltransferase genes involved in sinapate metabolism and lignin synthesis in Arabidopsis. J Biol Chem 276:4344–4349
Manavalan LP, Guttikonda SK, Tran LSP, Nguyen HT (2009) Physiological and molecular approaches to improve drought resistance in soybean. Plant Cell Physiol 50:1260–1276
Maruyama N, Salleh MRM, Takahashi K, Yagasaki K, Goto H, Hontani N (2002) Structure-physicochemical function relationships of soybean β-conglycinin heterotrimers. J Agric Food Chem 50:4323–4326
Meurer CA, Dinkins RD, Collins GB (1998) Factors affecting soybean cotyledonary node transformation. Plant Cell Rep 18:180–186
Nakamura I, Dube PH, Beachy Roger N (1993) Accumulation of the products of beta-conglycinin alpha’-subunit gene constitutively expressed in seeds and non-seed tissues of the transgenic Petunia plants. Plant Cell Physiol 34:865–872
Niu HS, Liu IM, Cheng JT, Lin CL, Hsu FL (2008) Hypoglycemic effect of syringin from Eleutherococcus senticosus in streptozotocin-induced diabetic rats. Planta Med 74:109–113
Oksman-Caldentey KM, Hiltunen R (1996) Transgenic crops for improved pharmaceutical products. Field Crops Res 45:57–69
Olhoft PM, Flagel LE, Donovan CM, Somers DA (2003) Efficient soybean transformation using hygromycin B selection in the cotyledonary-node method. Planta 216:723–735
Paz MM, Martinez JC, Kalvig AB, Fonger TM, Wang K (2006) Improved cotyledonary node method using an alternative explant derived from mature seed for efficient Agrobacterium-mediated soybean transformation. Plant Cell Rep 25:206–213
Ralph J, Lundquist K, Brunow G, Lu F, Kim H, Schatz PF, Marita JM, Hatfield RD, Christensen JH, Boerjan W (2004) Lignins: natural polymers from oxidative coupling of 4-hydroxyphenylpropanoids. Phytochem Rev 3:29–60
Ramakrishna A, Ravishankar GA (2011) Influence of abiotic stress signals on secondary metabolites in plants. Plant Signal Behav 6:1720–1731
Sakai T, Kogiso M (2008) Soy isoflavones and immunity. J Med Invest 55:167–173
Samac DA, Tesfaye M, Dornbusch M, Saruul P, Temple SJ (2004) A comparison of constitutive promoters for expression of transgenes in alfalfa (Medicago sativa). Transgenic Res 13:349–361
Sarah AO, Søren B, Birger LM (2009) Substrate specificity of plant UDP-dependent glycosyltransferases predicted from crystal structures and homology modeling. Phytochemistry 70:325–347
Savidge RA (1989) Coniferin, a biochemical indicator of commitment to tracheid differentiation in conifers. Can J Bot 67:2663–2668
Utsumi S, Matsumura Y, Mori T (1997) Structure–function relationships of soy proteins. In: Damodaran S, Paraf A (eds) Food proteins and their applications. Marcel Dekker, New York, pp 257–291
Verpoorte R, Memelink J (2002) Engineering secondary metabolite production in plants. Curr Opin Biotechnol 13:181–187
Verpoorte R, van der Heijden R, Memelink J (2000) Engineering the plant cell factory for secondary metabolite production. Transgenic Res 9:323–343
Yoshino M, Nagamatsu A, Tsutsumi K, Kanazawa A (2006) The regulatory function of the upstream sequence of the ββ-conglycinin α subunit gene in seed-specific transcription is associated with the presence of the RY sequence. Genes Genet Syst 81:135–141
Zakharov A, Giersberg M, Hosein F, Melzer M, Muntz K, Saalbach I (2004) Seed-specific promoters direct gene expression in non-seed tissue. J Exp Bot 55:1463–1471
Acknowledgements
This work was supported by the Next-Generation BioGreen 21 Program (Code PJ011150 and Code PJ011111022016 granted to Y.S. Chung and J. Nam, respectively), Rural Development Administration, and Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries (Code 112124-5 granted to Y.S. Chung), and National Research Foundation (NRF-2017R1C1B5017598 granted to T. Kwon) in Republic of Korea.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.

11816_2017_464_MOESM1_ESM.jpg
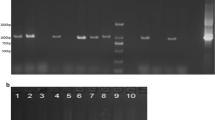
Verification of the introduced genes and expression levels in 35S-UGT72E3/2 transgenic plants (T0) using PCR and RT-PCR. a Genomic DNAs were extracted from T0 transgenic leaf tissues. Lane 1, UGT72E3/2 gene; Lane 2, Bar gene; Lane 3, DNA region between the 35S promoter and UGT72E3/2 gene; Lane 4, DNA region between the left border and Bar gene; Lane 5, DNA region between UGT72E3/2 gene and right border. b Total RNA was extracted from T0 transgenic leaf tissues. The TUB gene was used as a normalization control for leaf RNA levels. NT, non-transgenic plant; #1-#10, 35S-UGT72E3/2 transgenic lines (T0) (JPG 145 KB)
Rights and permissions
About this article
Cite this article
Kwon, T., Kim, H.J., Yun, S.Y. et al. Enhancement of syringin contents in soybean seeds with seed-specific expression of a chimeric UGT72E3/E2 gene. Plant Biotechnol Rep 11, 439–447 (2017). https://doi.org/10.1007/s11816-017-0464-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11816-017-0464-5