Abstract
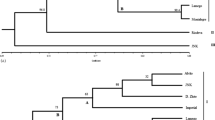
Germplasm characterization and evolutionary process in viable populations are important links between the conservation and utilization of plant genetic resources. Here, an investigation is made, based on molecular and biochemical techniques for assessing and exploiting the genetic variability in germplasm characterization of taro, which would be useful in plant breeding and ex situ conservation of taro plant genetic resources. Geographical differentiation and phylogenetic relationships of Indian taro, Colocasia esculenta (L.) Schott, were analyzed by random amplified polymorphic DNA (RAPD) and isozyme of seven enzyme systems with specific reference to the Muktakeshi accession, which has been to be proved resistant to taro leaf blight caused by P. colocasiae. The significant differentiations in Indian taro cultivars were clearly demonstrated by RAPD and isozyme analysis. RAPD markers showed higher values for genetic differentiation among taro cultivars and lower coefficient of variation than those obtained from isozymes. Genetic differentiation was evident in the taro accessions collected from different regions of India. It appears that when taro cultivation was introduced to a new area, only a small fraction of genetic variability in heterogeneous taro populations was transferred, possibly causing random differentiation among locally adapted taro populations. The selected primers will be useful for future genetic analysis and provide taro breeders with a genetic basis for selection of parents for crop improvement. Polymorphic markers identified in the DNA fingerprinting study will be useful for screening a segregating population, which is being generated in our laboratory aimed at developing a taro genetic linkage map.


Similar content being viewed by others
References
Bartish IV, Garkava LP, Rumpunen K, Nybom H (2000) Phylognetic relationship and differentiation among and within populations of Chaenomeles Lindl. (Rosaceae) estimated with RAPDs and isozymes. Theor Appl Genet 101:554–563
Brummer EC, Bouton JH, Korchert G (1995) Analysis of annual Medicago species using RAPD markers. Genome 38:362–367
Chalmers KJ, Waugh R, Sprent JI, Simmons AJ, Powell W (1992) Detection of genetic variation between and within populations of Gliricidi sepium and G. maculata using RAPD markers. Heredity 69:465–472
Garkava LP, Rumpunen K, Bartish IV (2000) Genetic relationships in Chaenomeles (Rosaceae) revealed by isozyme analysis. Sci Hortic 85:21–35
Hamrick JL, Allard RW (1972) Microgeographical variation in allozyme frequencies in Avena barbata. Proc Natl Acad Sci USA 69:2100–2104
Hamrick JL, Godt JW (1990) Allozyme diversity in plant species. In: Brown HD, Cleg MT, Kahler AL, Weir BS (eds) Plant population genetics, breeding, and genetics resources. Sinauer, Sunderland
Hedrick PW (2000) Genetics of populations, 2nd edn. Jones & Bartlett, Sudbury
Iglesias L, Lima H, Simon JP (1974) Isozyme identification of zygotic and nucellar seedlings in Citrus. J Heredity 65:81–84
Jackson GVH, Gollifer DE, Newhook FJ (1980) Studies on the taro leaf blight fungus Phytophthora colocasiae in the Solomon Islands: control by fungicides and spacing. Ann Appl Biol 96:1–10
Lebot V, Herail C, Gunua T, Pardales J, Prana M, Thongjiem M, Viet N (2003) Isozyme and RAPD variation among Phytophthora colocasiae isolates from South East Asia and the Pacific. Plant Pathol 52:303–313
Matos M, Pinto-Carnide O, Benito C (2001) Phylognetic relationships among Portuguese rye based on Isozyme, RAPD and ISSR markers. Hereditas 134:229–236
Messmer MM, Melchinger AE, Woodman WL, Lee EA, Lamkey KR (1991) Genetic diversity among progenitors and elite lines from the Iowa Stiff Stalk Synthetic (BSSS) maize population: comparison of allozyme and RFLP data. Theor Appl Genet 83:97–107
Misra RS, Chowdhury SR (1997) Phytophthora leaf blight disease in taro. Technical Bulletin Series 21, C.T.C.R.I. (ICAR), Trivandrum.
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA 70:3321–3323
Nevo E, Apelbaum-Elkaher I, Garty J, Beiles A (1997) Natural selection causes microscale allozyme diversity in wild barley and a lichen at ‘Evolution Canyon’, Mt. Carmel, Israel. Heredity 78:373–382
Ochiai T, Nguyen VX, Tahara M, Yoshino H (2001) Geographical differentiation of Asian taro, Colocasia esculanta (L.) Schott, detected by RAPD and Isozyme analyses. Euphytica 122:219–234
Selander RK, Korhonen TK, Vaisanen-Rhen V, Pattison PE, Caugant DA (1986) Genetic relationships and clonal structure of strains of Escherichia coli causing neonatal septicemia and meningitis. Infect Immun 52:213–222
Sharma K, Mishra AK, Misra RS (2008) A simple and efficient method for extraction of genomic DNA from tropical tuber crops. Afr J Biotechnol 7(8):1018–1022
Smith OS, Smith JSC, Bowen SL, Tenborg RA (1992) Numbers of RFLP probes necessary to show associations between lines. Maize Genet Coop News Lett 66:66
Soltis PS, Soltis D, Doyle J (1992) Molecular systematics of plants. Chapman and Hall, New York
Swoboda I, Bhalla PI (1997) RAPD analysis of genetic variation in the Australian fan flower, Scaevola. Genome 40:600–606
Thankappan M (1985) Leaf blight of taro-a review. J Root Crops 11:223–236
Tinker NA, Fortin MG, Mather DE (1993) Random amplified polymorphic DNA and pedigree relationships in spring barley. Theor Appl Genet 85:976–984
Wachira FN, Waugh R, Hachett CA, Powell W (1995) Detection of genetic diversity in tea (Camellia sinesis) using RAPD markers. Genome 38:201–210
Weir BS (1990) Genetic data analysis. Sinauer, Sunderland
Weir BS, Cockerham CC (1984) Estimating F. statistics for the analysis of population’s structure. Evolution 38:1358–1370
Wendel JF, Doyle JJ (1998) Phylogenetic incongruence: window into the genome history and molecular evolution. In: Soltis DE, Soltis PS, Doyle JJ (eds) Molecular systematics of Plants II. DNA Sequenceing. Kluwer, Boston, pp 265–296
Whitkus R, Doebley J, Wendel JF (1994) DNA-based markers in plants. In: Philllips L, Vasil IK (eds) Nuclear DNA markers in systematics and evolution. Kluwer, Amsterdam, pp 116–141
Wolff K, Peters-van Rijn J (1993) Rapid detection of genetic variability in chrysanthemum (Dendanthema grandiflora Tzvelev) using random primers. Heredity 71:335–341
Yeh FC, Yang R (1999) Microsoft window-based freeware for population genetic analysis (POPGENE version 1.31). University of Alberta, Canada
Acknowledgments
The funding provided for conducting the research work by the Indian Council of Agricultural Research, New Delhi, is gratefully acknowledged. The authors thank Director, Central Tuber Crops Research Institute, Thiruvananthapuram, for providing the infrastructure facilities.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sharma, K., Mishra, A.K. & Misra, R.S. The genetic structure of taro: a comparison of RAPD and isozyme markers. Plant Biotechnol Rep 2, 191–198 (2008). https://doi.org/10.1007/s11816-008-0061-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11816-008-0061-8