Abstract
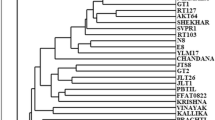
Indian jujube germplasm characterization, evaluation and improvement are fundamentally based on morpho-physiological traits. The lack of break through has been due to under utilization of genetic variability for superior quality and high yield potential. We report the extent of genetic diversity and relationships among 22 putative Ziziphus mauritiana varieties using nuclear ribosomal DNA and RAPD polymorphism. A hybrid protocol of DNA isolation was developed to obtain high molecular weight DNA. Ten RAPD primers generated 101 markers and detected intra-specific variation amounting to 85.15 % polymorphism in banding patterns delineating them into four main clusters and variety Tikadi as a distinct out group. Besides, internal transcribed spacer (ITS) length variations, single nucleotide polymorphism and insertions/deletions (INDELS) in nuclear rDNA region delineated test varieties and revealed lineages with GenBank reference sequences of Z. mauritiana. The novel gene sequences exhibiting nucleotide variations have been submitted to NCBI database and assigned accession numbers from JQ627029 to JQ627050. Higher values of Nei’s genetic diversity (h), Shannon information index (i) and genetic distance analysis validate higher genetic diversity among Indian jujube varieties. The genetically distinct varieties of different phylogenetic clades of RAPD and ITS phylogram can be effectively utilized as parents for crop improvement.





Similar content being viewed by others
References
Andrew M, Kohn LM (2009) Single nucleotide polymorphism based diagnostic system for crop-associated Sclerotinia species. Appl Environ Microbiol 75:5600–5606
Awasthi OP, More TA (2009) Genetic diversity and status of Ziziphus in India. Acta Hortic 840:33–40
Bhatia A, Mishra T (2009) Free radical scavenging and antioxidant potential of Ziziphus mauritiana (Lamk.) seed extract. J Complement Integr Med 8:42–46
Carvalho A et al (2009) Genetic diversity among old Portugese bread wheat cultivars and Botanical varieties evaluated by ITS rDNA PCR–RFLP markers. J Genet 88(3):363–367
Dahiru D, Obidoa O (2007) Pretreatment of albino rats with aqueous leaf extract of Ziziphus mauritiana protects against alcohol induced liver damage. Trop J Pharm Res 6:705–710
Devanshi et al (2007) Molecular profiling and genetic relationship among ber (Ziziphus sp.) genotypes using RAPD markers. Indian J Genet 67(2):121–127
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12(1):13
El-Mouei R et al (2011) Characterization and estimation of genetic diversity in Citrus rootstocks. Int J Agric Biol 13:571–575
Fu JX et al (2007) Identification and classification of ber cultivars based on ISSR and RAPD analysis. Acta Hortic 764:119–126
Godara NR (1980) Studies on floral biology and compatibility behavior in ber (Ziziphus mauritiana Lamk.) Ph.D. thesis. Haryana Agricultural University, Hisar
Gupta RB et al (2003) Study on vegetative characters of some cultivated and wild forms of ber (Ziziphus spp.). Haryana J Hortic Sci 32(1/2):15–18
Hershkovitz MA et al (1999) Ribosomal DNA sequences and angiosperm systematics. In: Hollingsworth PM, Bateman RM, Gornall RJ (eds) Molecular systematics and plant evolution. Taylor and Francis, London, pp 268–326
Hillis DM, Bull JJ (1993) An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analyses. Syst Biol 42:182–192
Hsiao C et al (1994) Phylogenetic relationships of 10 grass species: an assessment of phylogenetic utility of the internal transcribed spacer region in nuclear ribosomal DNA in monocots. Genome 37:112–120
Hudina M et al (2008) Phenolic compounds in the fruit of different varieties of Chinese jujube (Ziziphus jujuba Mill.). J Hortic Sci Biotechnol 83:305–308
Islam MB, Simmons MP (2006) A thorny dilemma: testing alternative intra-generic classifications within Ziziphus (Rhamnaceae). Syst Bot 31(4):826–842
Jorgenson RD, Cluster PD (1988) Modes and tempos in the evolution of nuclear ribosomal DNA: new characters of evolutionary studies and new markers for genetic and population studies. Ann Mo Bot Gard 75:1238–1247
Kakani RK et al (2011) Assessment of genetic diversity in Trigonella foenum-graecum based on nuclear ribosomal DNA, internal transcribed spacer and RAPD analysis. Plant Mol Biol Rep 29:315–323
Lewontin RC (1972) The apportionment of human diversity. Evol Biol 6:391–398
Li L et al (2009) Studies on the phylogenetic relationships of Chinese Ziziphus by RAPD technique. Acta Hortic Sin 36(4):475–480
Liu MJ, Cheng CY (1995) A taxonomy study on the genus Ziziphus. Acta Hortic 390:161–165
Liu P et al (2006) Genetic diversity evaluation of Ziziphus jujuba cv. Zanhuangdazao. J Fruit Sci 23(5):685–689
Ma QH et al (2011) Development and characterization of SSR markers in Chinese jujube (Ziziphus jujuba Mill.) and its related species. Sci Hortic 129:597–602
Martins SR et al (2006) RAPD analysis of genetic diversity among and within Portuguese landraces of common white bean (Phaseolus vulgaris L.). Sci Hortic 108:133–142
Mishra T et al (2011) Anticancer potential of aqueous ethanol seed extract of Ziziphus mauritiana against cancer cell lines and ehrlich ascites carcinoma. Evid Complement Altern Med. doi:10.1155/2011/765029
Morton J (1987) Indian jujube. In: Mortan JF, Miami FL (eds) Fruits of warm climates. Center for New Crops & Plant Products, Purdue University, Lafayette, IN, USA, pp 272–275. http://www.hort.purdue.edu/newcrop/morton/indianjujube.html
Morton JF (2005) Indian jujube. J Exp Bot 56(422):3082–3092
Navjot et al (2009) Genetic parameters, character association and path analysis for fruit yield and its components in ber (Ziziphus mauritiana). Indian J Agric Sci 79(12):1000–1002
Ndhala AR et al (2006) Antioxidant potentials and degree of polymerization of six wild fruits. Life Sci Res Assays 1:87–92
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA 70:3321–3323
Obeed RS et al (2008) Fruit properties and genetic diversity of five ber (Ziziphus mauritiana Lamk.) cultivars. Pak J Biol Sci 11(6):888–893
Pareek OP (2001) Fruits for the future 2: ber. International Centre for Underutilized Crop, Redwood Books, Wiltshire, p 290
Pareek OP, Nath V (1996) Ber. Coordinated fruit research in Indian arid zone—a two decades profiles (1976–1995). National Research Centre for Arid Horticulture, Bikaner, pp 9–30
Pathak R et al (2011) Assessment of genetic diversity in clusterbean using nuclear rDNA and RAPD markers. J Food Legumes 24(3):180–183
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population software for teaching and research. Mol Ecol Notes 6:288–295
Perrière G, Gouy M (1996) WWW-Query: an on-line retrieval system for biological sequence banks. Biochimie 78:364–369
Qi J et al (2009) Construction of a dense genetic linkage map and QTL analysis of trunk diameter in Chinese jujube. Scientia Silvae Sinicae 45(8):44–49
Raturi A et al (2012) Molecular characterization of Vigna radiata (L.) Wilczek genotypes based on nuclear ribosomal DNA and RAPD polymorphism. Mol Biol Rep 39(3):2455–2465
Raut GR et al (2010) Study of relationships among twelve Phyllanthus species with the use of molecular markers. Czech J Genet Plant Breed 46:135–141
Rohlf FJ (1997) NTSYS pc: numerical taxonomy and multivariate analysis system Version 2.02h. Exeter software, New York
Saran PL et al (2006) Characterization of ber (Ziziphus mauritiana Lamk.) genotypes. Haryana J Hortic Sci 35(3/4):215–218
Sarkhosh A et al (2009) Evaluation of genetic diversity among Iranian soft-seed pomegranate accessions by fruit characteristics and RAPD markers. Sci Hortic 121:313–319
Sehrawat SK et al (2006) DNA finger printing of ber (Ziziphus mauritiana Lamk) germplasm. Haryana J Hortic Sci 35(3/4):213–214
Singh RS et al (1998) Micrometeorology of ber (Ziziphus mauritiana Lamk.) orchard grown under rainfed arid conditions. Indian J Hortic 55(2):97–107
Singh SK et al (2005) Morel Phylogeny and diagnostics based on restriction fragment length polymorphism analysis of ITS region of 5.8S ribosomal DNA. J Plant Biochem Biotechnol 14:179–183
Singh AK et al (2006) Genetic diversity in ber (Ziziphus sp.) revealed by AFLP markers. J Hortic Sci Biotechnol 81:205–210
Singh SK et al (2011) Molecular assessment of inter-specific genetic diversity in selected species of Prosopis revealed by RAPD. Indian J Agric Sci 81(2):167–171
Sneath PHA, Sokal RR (1973) Numerical taxonomy. W. H. Freeman & Co., San Francisco, CA
Souza IGB et al (2011) RAPD analysis of the genetic diversity of mango (Mangifera indica) germplasm in Brazil. Genet Mol Res 10(4):3080–3089
Vashishtha BB (2001) Ber varieties: a monograph. Agrobios (India), Jodhpur, p 97
Verheij EWM, Calabura M (1991) Plant resources of South-East Asia 2. In: Verheij EWM, Coronel RE (eds) Edible fruits and nuts. PROSEA, Pudoc, Wageningen, pp 223–225
Wang YK et al (2009) Conservation, characterization, evaluation and utilization of Chinese jujube germplasm resources. Acta Hortic 840:235–240
Wen YF et al (2009) AFLP analysis of genetic relationship and discrimination of Chinese jujube germplasm resources. Acta Hortic 840:117–124
White TJ et al (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innes MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols, a guide to methods and applications. Academic Press, San Diego, pp 315–322
Williams JGK et al (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18:6231–6235
Xiao LQ et al (2010) High nrDNA ITS polymorphism in the ancient extant seed plant Cycas: incomplete concerted evolution and the origin of pseudogenes. Mol Phylogenet Evol 55:168–177
Acknowledgments
The authors are thankful to Dr. M. M. Roy, Director, CAZRI, Jodhpur for providing necessary laboratory and field facilities and to Dr. O. P. Pareek and Dr. B. B. Vashishtha for developing Z. mauritiana varieties and nurturing orchard at CR Farm CAZRI, Jodhpur, Rajasthan.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Singh, S.K., Meghwal, P.R., Pathak, R. et al. Assessment of Genetic Diversity Among Indian Jujube Varieties Based on Nuclear Ribosomal DNA and RAPD Polymorphism. Agric Res 3, 218–228 (2014). https://doi.org/10.1007/s40003-014-0112-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40003-014-0112-z