Abstract
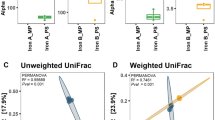
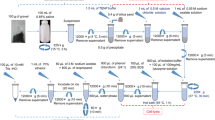
Extraction of high-quality microbial DNA from contaminated environmental samples is an essential step in microbial ecological study. Based on previously published methods for soil and sediment samples, a modified pretreatment method was developed for extracting microbial DNA from heavily contaminated river sediment samples via selection of optimal pretreatment parameters (i.e., reagent solution, reaction duration, and temperature). The pretreatment procedure involves washing the river sediment sample for three times with a solution containing 0.1 mol·L−1 ethylene diamine tetraacetic acid (EDTA), 0.1 mol·L−1 Tris (pH 8.0), 1.5 mol·L−1 NaCl, 0.1 mol·L−1 NaH2PO4, and Na2HPO4 at 65°C with 180 r·min−1 for 15 min to remove humic materials and heavy metals prior to the employment of standard DNA extraction procedures. We compared the results of standard procedure DNA extraction following pretreatment, without pretreatment, and with using a commercial PowerSoil™ DNA Isolation Kit. The results indicated that the pretreatment significantly improved the DNA quality based on DNA yield, DNA fragment length, and determination of prokaryotic diversity. Prokaryotic diversity exhibited in the DNA with the pretreatment was also considerably higher than that extracted with the PowerSoil™ DNA Isolation Kit only. The pretreatment method worked well even with a small amount of sediment sample (0.25 g or even lower). The method provides a novel, simple, cost-effective tool for DNA extraction for microbial community analysis in environmental monitoring and remediation processes.
Similar content being viewed by others
References
Cai J N, Cao Y Z, Tan H J, Wang Y M, Luo J Q. Fractionation and ecological risk of metals in urban river sediments in Zhongshan City, Pearl River Delta. Journal of Environmental Monitoring, 2011, 13(9): 2450–2456
Mehler W T, Li H Z, Lydy M J, You J. Identifying the causes of sediment-associated toxicity in urban waterways of the Pearl River Delta, China. Environmental Science & Technology, 2011, 45(5): 1812–1819
Santhiya G, Lakshumanan C, Selvin J, Asha D. Microbiological analysis of seawater and sediments in urban shorelines: occurrence of heavy metals resistance bacteria on Chennai beaches, Bay of Bengal. Microchemical Journal, 2011, 99(2): 197–202
Wu L, Kellogg L, Devol A H, Tiedje J M, Zhou J. Microarray-based characterization of microbial community functional structure and heterogeneity in marine sediments from the Gulf of Mexico. Applied and Environmental Microbiology, 2008, 74(14): 4516–4529
Pires A C C, Cleary D F R, Almeida A, Cunha Â, Dealtry S, Mendonça-Hagler L C S, Smalla K, Gomes N C M. Denaturing gradient gel electrophoresis and barcoded pyrosequencing reveal unprecedented archaeal diversity in mangrove sediment and rhizosphere samples. Applied and Environmental Microbiology, 2012, 78(16): 5520–5528
Hollister E B, Engledow A S, Hammett A J M, Provin T L, Wilkinson H H, Gentry T J. Shifts in microbial community structure along an ecological gradient of hypersaline soils and sediments. ISME Journal, 2010, 4(6): 829–838
Bowman J S, Rasmussen S, Blom N, Deming J W, Rysgaard S, Sicheritz-Ponten T. Microbial community structure of Arctic multiyear sea ice and surface seawater by 454 sequencing of the 16S RNA gene. The ISME Journal, 2012, 6(1): 11–20
Ning J, Liebich J, Kästner M, Zhou J, Schäffer A, Burauel P. Different influences of DNA purity indices and quantity on PCR-based DGGE and functional gene microarray in soil microbial community study. Applied Microbiology and Biotechnology, 2009, 82(5): 983–993
Delmont T O, Robe P, Clark I, Simonet P, Vogel TM. Metagenomic comparison of direct and indirect soil DNA extraction approaches. Journal of Microbiological Methods, 2011, 86(3): 397–400
Amorim J H, Macena T N S, Lacerda-Junior G V, Rezende R P, Dias J C T, Brendel M, Cascardo J C M. An improved extraction protocol for metagenomic DNA from a soil of the Brazilian Atlantic Rainforest. Genetics and Molecular Research, 2008, 7(4): 1226–1232
Fitzpatrick K A, Kersh G J, Massung R F. Practical method for extraction of PCR-quality DNA from environmental soil samples. Applied and Environmental Microbiology, 2010, 76(13): 4571–4573
Fortin N, Beaumier D, Lee K, Greer C W. Soil washing improves the recovery of total community DNA from polluted and high organic content sediments. Journal of Microbiological Methods, 2004, 56(2): 181–191
Purohit H J, Kapley A, Moharikar A A, Narde G. A novel approach for extraction of PCR-compatible DNA from activated sludge samples collected from different biological effluent treatment plants. Journal of Microbiological Methods, 2003, 52(3): 315–323
Sagova-Mareckova M, Cermak L, Novotna J, Plhackova K, Forstova J, Kopecky J. Innovative methods for soil DNA purification tested in soils with widely differing characteristics. Applied and Environmental Microbiology, 2008, 74(9): 2902–2907
Leuko S, Goh F, Ibáñez-Peral R, Burns B P, Walter M R, Neilan B A. Lysis efficiency of standard DNA extraction methods for Halococcus spp. in an organic rich environment. Extremophiles, 2008, 12(2): 301–308
Zhou J, Bruns M A, Tiedje J M. DNA recovery from soils of diverse composition. Applied and Environmental Microbiology, 1996, 62(2): 316–322
Braida M D, Daniels L M, Kitts C L. Removal of PCR inhibitors from soil DNA by chemical flocculation. Journal of Microbiological Methods, 2003, 52(3): 383–393
Leff L G, Dana J R, McArthur J V, Shimkets L J. Comparison of methods of DNA extraction from stream sediments. Applied and Environmental Microbiology, 1995, 61(3): 1141–1143
Miller D N, Bryant J E, Madsen E L, Ghiorse W C. Evaluation and optimization of DNA extraction and purification procedures for soil and sediment samples. Applied and Environmental Microbiology, 1999, 65(11): 4715–4724
Rojas-Herrera R, Narváez-Zapata J, Zamudio-Maya M, Mena-Martínez ME. A simple silica-based method for metagenomic DNA extraction from soil and sediments. Molecular Biotechnology, 2008, 40(1): 13–17
Lakay F M, Botha A, Prior B A. Comparative analysis of environmental DNA extraction and purification methods from different humic acid-rich soils. Journal of Applied Microbiology, 2007, 102(1): 265–273
Liu J, Deng D Y, Xu M Y, Sun G P. Characteristics of organic pollutants in the sediments from a typical electronics industrial zone. Environmental Sciences (China), 2013, 34(3): 326–333 (in Chinese)
Deng D Y, Sun G P, Guo J, Zhang H T, Zhang Q, Xu M Y. Investigation on the distribution and potential ecological risk of heavy metal in the sediments from typical electrical industrial zone. Environmental Sciences (China), 2012, 33(5): 1700–1766 (in Chinese)
Fu J, Mai B, Sheng G, Zhang G, Wang X, Peng P, Xiao X, Ran R, Cheng F, Peng X, Wang Z, Wa Tang U. Persistent organic pollutants in environment of the Pearl River Delta, China: an overview. Chemosphere, 2003, 52(9): 1411–1422
Enright M J, Scott E E, Chang K. Regional Powerhouse: The Greater Pearl River Delta and the Rise of China. Chichester: Wiley, 2005
Csaikl U, Bastian H, Brettschneider R, Gauch S, Meir A, Schauerte M, Scholz F, Sperisen C, Vornam B, Ziegenhagen B. Comparative analysis of different DNA extraction protocols: a fast, universal maxi-preparation of high quality plant DNA for genetic evaluation and phylogenetic studies. Plant Molecular Biology Reporter, 1998, 16(1): 69–86
Watson R J, Blackwell B. Purification and characterization of a common soil component which inhibits the polymerase chain reaction. Canadian Journal of Microbiology, 2000, 46(7): 633–642
Lahiri D K, Schnabel B. DNA isolation by a rapid method from human blood samples: effects of MgCl2, EDTA, storage time, and temperature on DNA yield and quality. Biochemical Genetics, 1993, 31(7–8): 321–328
Wang G C, Wang Y. Frequency of formation of chimeric molecules as a consequence of PCR coamplification of 16S rRNA genes from mixed bacterial genomes. Applied and Environmental Microbiology, 1997, 63(12): 4645–4650
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Fang, Y., Xu, M., Chen, X. et al. Modified pretreatment method for total microbial DNA extraction from contaminated river sediment. Front. Environ. Sci. Eng. 9, 444–452 (2015). https://doi.org/10.1007/s11783-014-0679-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11783-014-0679-4