Abstract
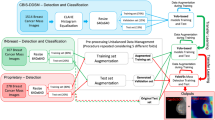
Active contours model (ACM) has been extensively used in computer vision and image processing. In recent studies, convolutional neural networks (CNNs) have been combined with ACM replacing the user in the process of contour evolution and image segmentation to eliminate limitations associated with ACM dependence on energy functional parameters and initialization. However, prior studies did not aim for automatic initialization, which is addressed in this article. In addition to manual initialization, current methods are highly sensitive to the initial location and fail to delineate borders accurately. We propose a fully automatic image segmentation method to address problems of manual initialization, insufficient capture range, and poor convergence to boundaries, in addition to the problem of assignment of energy functional parameters. We train two CNNs, one of which generating ACM weighting parameters and the other generating a ground truth mask to extract distance transform (DT) and an initialization circle. DT is used to form a vector field pointing from each pixel of the image towards the closest ground truth boundary point. Vector magnitudes are equal to the Euclidean distance between each pixel and the closest ground truth boundary point. We evaluate our method on four publicly available datasets, including two building instance segmentation datasets, i.e., Vaihingen and Bing huts, and two mammography image datasets, INBreast and DDSM-BCRP. Our approach achieves state-of-the-art results in mean Intersection over Union (mIoU), Dice similarity coefficient and Boundary F-score (BoundF) with the values of 92.33%, 92.44%, and 86.57% for Vaihingen dataset, and 87.12%, 86.86%, and 66.91% for Bing huts dataset. We obtained the Dice similarity coefficient values of 94.23% and 90.89% for the INBreast and DDSM-BCRP, respectively.








Similar content being viewed by others
References
Kass, M., Witkin, A., Terzopoulos, D.: Snakes: active contour models. Int. J. Comput. Vis. 1(4), 321–331 (1988). https://doi.org/10.1007/BF00133570
Cohen, L.D., Cohen, I.: Finite-element methods for active contour models and balloons for 2-D and 3-D images. IEEE Trans. Pattern Anal. Mach. Intell. 15(11), 1131–1147 (1993). https://doi.org/10.1109/34.244675
Xu, C., Prince, J.L.: Snakes, shapes, and gradient vector flow. IEEE Trans. Image Process. 7(3), 359–369 (1998). https://doi.org/10.1109/83.661186
Sum, K.W., Cheung, P.Y.S.: Boundary vector field for parametric active contours. Pattern Recognit. 40(6), 1635–1645 (2007). https://doi.org/10.1016/j.patcog.2006.11.006
Xie, X., Mirmehdi, M.: MAC: magnetostatic active contour model. IEEE Trans. Pattern Anal. Mach. Intell. 30(4), 632–646 (2008). https://doi.org/10.1109/TPAMI.2007.70737
Wang, T., Cheng, I., Basu, A.: Fluid vector flow and applications in brain tumor segmentation. IEEE Trans. Biomed. Eng. 56(3), 781–789 (2009). https://doi.org/10.1109/TBME.2009.2012423
Cohen, L.D.: On active contour models and balloons. CVGIP Image Underst. 53(2), 211–218 (1991). https://doi.org/10.1016/1049-9660(91)90028-N
Rupprecht, C., Huaroc Moquillaza, E., Baust, M., Navab, N.: Deep active contours (2016). arXiv preprint arXiv:160705074
Marcos, D., Tuia, D., Kellenberger, B., Zhang, L., Bai, M., Liao, R., Urtasun, R.: Learning deep structured active contours end-to-end. In: 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pp. 8877–8885 (2018). https://doi.org/10.1109/CVPR.2018.00925
Cheng, D., Liao, R., Fidler, S., Urtasun, R.: Darnet: deep active ray network for building segmentation. In: 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pp. 7431–7439 (2019). https://doi.org/10.1109/CVPR.2019.00761
Gur, S., Shaharabany, T., Wolf, L.: End to end trainable active contours via differentiable rendering (2019). arXiv preprint arXiv:191200367
Hoogi, A., Subramaniam, A., Veerapaneni, R., Rubin, D.L.: Adaptive estimation of active contour parameters using convolutional neural networks and texture analysis. IEEE Trans. Med. Imaging 36(3), 781–791 (2017). https://doi.org/10.1109/TMI.2016.2628084
Le, T.H.N., Quach, K.G., Luu, K., Duong, C.N., Savvides, M.: Reformulating level sets as deep recurrent neural network approach to semantic segmentation. IEEE Trans. Image Process. 27(5), 2393–2407 (2018). https://doi.org/10.1109/TIP.2018.2794205
Albalooshi, F.A., Sidike, P., Sagan, V., Albalooshi, Y., Asari, V.K.: Deep belief active contours (DBAC) with its application to oil spill segmentation from remotely sensed sea surface imagery. Photogram. Eng. Remote Sens. 84(7), 451–458 (2018). https://doi.org/10.14358/PERS.84.7.451
Hatamizadeh, A., Sengupta, D., Terzopoulos, D.: End-to-end deep convolutional active contours for image segmentation (2019). arXiv preprint arXiv:190913359
Wang, Z., Acuna, D., Ling, H., Kar, A., Fidler, S.: Object instance annotation with deep extreme level set evolution. In: 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pp. 7500–7508 (2019). https://doi.org/10.1109/CVPR.2019.00768
Hatamizadeh, A., Sengupta, D., Terzopoulos, D.: End-to-end trainable deep active contour models for automated image segmentation: delineating buildings in aerial imagery. In: European Conference on Computer Vision, vol. 12357, pp. 730–746. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-58610-2_43
2d semantic labeling contest. In: International Society for Photogrammetry and Remote Sensing. http://www2.isprs.org/commissions/comm3/wg4/semantic-labeling.html
Moreira, I.C., Amaral, I., Domingues, I., Cardoso, A., Cardoso, M.J., Cardoso, J.S.: Inbreast: toward a full-field digital mammographic database. Acad. Radiol. 19(2), 236–248 (2012). https://doi.org/10.1016/j.acra.2011.09.014
Heath, M., Bowyer, K., Kopans, D., Kegelmeyer, P., Moore, R., Chang, K., Munishkumaran, S.: Current status of the digital database for screening mammography. In: Karssemeijer, N., Thijssen, M., Hendriks, J., van Erning, L. (eds.) Digital Mammography, vol. 13, pp. 457–460. Springer, Dordrecht (1998). https://doi.org/10.1007/978-94-011-5318-8_75
Ball, J.E., Bruce, L.M.: Digital mammographic computer aided diagnosis (cad) using adaptive level set segmentation. In: 2007 29th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, pp. 4973–4978 (2007). https://doi.org/10.1109/IEMBS.2007.4353457
Zhu, W., Xiang, X., Tran, T.D., Hager, G.D., Xie, X.: Adversarial deep structured nets for mass segmentation from mammograms. In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), pp. 847–850 (2018). https://doi.org/10.1109/ISBI.2018.8363704
Li, H., Chen, D., Nailon, W.H., Davies, M.E., Laurenson, D.: Improved breast mass segmentation in mammograms with conditional residual u-net. In: Stoyanov, D., Taylor, Z., Kainz, B., Maicas, G., Beichel, R.R., Martel, A., Maier-Hein, L., Bhatia, K., Vercauteren, T., Oktay, O., Carneiro, G., Bradley, A.P., Nascimento, J., Min, H., Brown, M.S., Jacobs, C., Lassen-Schmidt, B., Mori, K., Petersen, J., Estépar, R.S.J., Schmidt-Richberg, A., Veiga, C. (eds.) Image Analysis for Moving Organ, Breast, and Thoracic Images, vol. 11040, pp. 81–89. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00946-5
Singh, V.K., Rashwan, H.A., Romani, S., Akram, F., Pandey, N., Sarker, M.K., Saleh, A., Arenas, M., Arquez, M., Puig, D., Torrents-barrena, J.: Breast tumor segmentation and shape classification in mammograms using generative adversarial and convolutional neural network. Expert Syst. Appl. 139, 1–14 (2020). https://doi.org/10.1016/j.eswa.2019.112855
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
This articles does not contain data from participants.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Akbarimoghaddam, P., Ziaei, A. & Azarnoush, H. Deep active contours using locally controlled distance vector flow. SIViP 16, 1773–1781 (2022). https://doi.org/10.1007/s11760-022-02134-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11760-022-02134-1