Abstract
The surveillance of sewage water has become an extremely essential tool to trace the circulation of viruses in a population and to predict the outbreak of viral diseases. Sewage monitoring is important for those viruses which cause subclinical infections since it is difficult to determine their prevalence. Polyomaviruses are ubiquitously present, circular double-stranded DNA viruses that can infect humans as well. Among all human polyomaviruses, BK polyomavirus and JC polyomavirus associated with the development of aggressive diseases in immunocompromised individuals, are highly prevalent. This study aimed to investigate the presence and the quantitative prevalence of these two disease-associated human polyomaviruses in sewage water collected from six drains of Lahore, Pakistan. The viruses present in the environmental samples were concentrated by PEG method before isolating viral nucleic acids. Conventional PCR amplifications were performed for molecular detection of BK polyomavirus and JC polyomavirus targeting their large tumor antigen genetic region. The presence of BK polyomavirus and JC polyomavirus was confirmed in the DNA extracted from concentrated sewage samples of each drain by performing both qualitative and quantitative PCR. Our data shows that the viral load ranged from 1278 to 178368 copies per µg of environmental DNA for BK polyomavirus and 5173 to 79129 copies per µg of environmental DNA for JC polyomavirus. In conclusion, here we report first time the detection of BK polyomavirus and JC polyomavirus in sewage water collected from six main drains in urban areas of Lahore, Pakistan showing the high prevalence of these viruses in the Pakistani population. This assay could be used as a proxy to determine the prevalence of these viruses in the Pakistani population.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Polyomaviruses (PyVs) are ubiquitously present, circular double-stranded DNA viruses whose natural hosts are primarily birds and mammals (Nomburg et al. 2022). Studies have reported that PyVs also infect the human population (Kamminga et al. 2018) The family of polyomaviruses infecting humans is rapidly expanding with 14 different species of human polyomaviruses being discovered till date (Linthorst et al. 2022). Not all of the human polyomaviruses are associated with the development of diseases but only four human polyomaviruses are known to be associated with aggressive diseases in immunocompromised individuals (Condez et al. 2021). Of these, BK polyomaviruses (BKPyV) and JC polyomavirus (JCPyV) were the foremost human polyomaviruses to be described and to be linked with human malignancies.
BKPyV & JCPyV genomes have been found in brain cells, immune cells, tonsils, respiratory tract, blood mononuclear cells, lymph nodes, and kidneys (Ciotti et al. 2019). Both viruses cause asymptomatic primary infection at an early age and remain latent in renal tissues and B lymphocytes until the immune system is suppressed in immunocompetent individuals (Whiley et al. 2001). In immunocompromised individuals, under conditions including diabetes, organ transplantation, anti-tumor therapy, and pregnancy (Barbanti-Brodano et al. 1998; Limaye et al. 2001), both viruses become reactivated from latent infection to a lytic state of infection (Iaconelli et al. 2015). The reactivation of BKPyV and JCPyV is associated with the development of devastating human diseases including polyomavirus-associated nephropathy, hemorrhagic cystitis, and progressive multifocal leukoencephalopathy (Dehcheshmeh et al. 2020).
Globally, BKPyV and JCPyV are highly prevalent, infecting about 70% and more than 90% of the adult population, respectively (Bouvard et al. 2012). BKPyV and JCPyV are characterized as small non-enveloped viruses, which contain a single double-stranded DNA of approximately 5 kb enclosed in an icosahedral capsid. The genome of both viruses encompasses two transcriptional active regions separated by a hypervariable non-coding control region, harbouring an ori gene essential for replication and transcription of early and late regions. The early region of both polyomaviruses is involved in the expression of regulatory proteins “tumor antigens” (T-Ag), whereas the late region encodes capsid proteins (Van Ghelue et al. 2012). The early region of BKPyV is translated into three tumor antigens (T-Ag) including large tumor antigen (LT-Ag), small tumor antigen (ST-Ag), and truncated T antigen (Trunc T-Ag) whereas the JCPyV early region encodes some splice variants (T’135, T’136, and T’165) in addition to LT-Ag and ST-Ag (Khalili et al. 2008). The late region of BKPyV and JCPyV is translated into structural capsid proteins including viral protein 1 (VP 1), viral protein 2 (VP 2), and viral protein 3 (VP 3) as well as non-structural agnoprotein (De Gascun and Carr 2013).
Both BKPyV and JCPyV have been known to spread through the respiratory tract, blood transfusion, seminal fluid, and/or organ transplantation. However, some reports describing the shedding of these viruses through urine and feces (Condez et al. 2021), and subsequently their detection in the sewage samples suggest the possibility of transmission through the urine-oral or fecal–oral route (Dehcheshmeh et al. 2020; Condez et al. 2021). Recently, a few studies in developed countries have reported that up to 90% of the human population has been exposed to human polyomaviruses, with several infections occurring at childhood ages (Egli et al. 2009; Kean et al. 2009; Šroller et al. 2016). However, no research study has been performed about the occurrence of polyomaviruses and the potential health hazards they might be causing in the Pakistani population. Therefore, the purpose of the research project proposed here is to detect polyomaviruses in different sewage samples from the city of Lahore, by using molecular tools specifically targeting the detection of polyomaviruses. Such efforts to detect these highly important but previously neglected polyomaviruses, are important as they may yield valuable insights into the establishment of latent infections and viral carcinogenesis.
Material and methods
Collection of sewage samples and concentration of viruses
Sewage (1 L) samples were collected from different drains located in Lahore in sterile collection bottles. Adequate personal protective equipments were used by personnels collecting sewage samples. The collected sample was mixed properly before aliquoting and stored at -20 °C. To detect viruses in sewage water, viruses were concentrated by polyethylene glycol (PEG) method (Hjelmsø et al. 2017). Briefly, 25 ml of glycine buffer (0.05 M glycine, 3% beef extract, pH 9.6) was mixed with 200 ml of sewage water in a sterile beaker to release viral particles from organic materials and then subjected to centrifugation at 7000 rpm for 30 min at 4 °C. The collected supernatant was filtered through 0.45 µm polyethersulfone membrane (PES) to remove eukaryotic and bacterial cells. The pH of the filtrate was checked and adjusted to 3.7 with 6 M HCl. Both PEG 8000 (80 g/L) and NaCl (17.5 g/L) were added to the filtrate and then incubated in a shaker at 4 °C for 15–20 h. The incubated filtrate was subjected to centrifugation at 7000 rpm for 2 h to precipitate viruses. The resulting pellet was dissolved in 1 ml of phosphate buffer saline (PBS) (0.137 M NaCl, 0.0027 M KCl, 0.01 M Na2HPO4, 0.0018 M KH2PO4, pH 7.4) and stored at -80 °C until further processing.
DNA extraction from sewage samples
The viral genome DNA was extracted from concentrated sewage samples by the TRIzol method. Primarily, 1 ml of TRIzol reagent was mixed with 400 µl of each concentrated sample following the addition of 300 µl of chloroform in sterile eppendorf tubes. The mixture was vortexed for 10 s, incubated at room temperature for 3 min, and centrifuged at 12,000 rpm for 10 min at 4 °C. After centrifugation, the middle layer was collected into a sterile eppendorf tube, mixed with equal volume of ice chilled isopropanol, and incubated at room temperature for 10 min following centrifugation. Pellet was washed with 70% ethanol, air-dried, and dissolved in 50 µl of DEPC treated water. The quality and quantity of isolated DNA were checked by gel electrophoresis and Nanodrop 2000.
Detection of BKPyV and JCPyV in sewage water by conventional PCR
The presence of BKPyV and JCPyV were detected in sewage water by using conventional PCR. PCRs were performed using the primer pairs specifically amplifying LT-Ag of BKPyV and JCPyV. A forward primer 5'-AATATTATGCCCAGCACACATG-'3 and a reverse primer 5'-CTTTCCCTCTGATCTACACCAG-'3 were used to amplify 171 bps of the Lt-Ag region of BKPyV. Similarly, a pair of forward primer 5'-AGAGTGTTGGGATCCTGTGTTT-'3 and reverse primer 5'-TTGCAGGGCATTTTGTTTTTTAC-'3 were used to amplify the 177 bps region of LT-Ag of JCPyV (Hussain et al. 2017). PCR was performed in a thermal cycler (applied B® 2720 thermal cycler) using the following optimized conditions: initial denaturation of DNA at 95 °C for 5 min followed by 35 cycles at 95 °C for 30 s, 57 °C for 45 s and 72 °C for 45 s and final extension at 72 °C for 10 min. Genes of LT-Ag of BKPyV and JCPyV separately cloned in pcDNA3 expression vectors, provided by Dr. Tommasino Lab, International Agency for Research on Cancer (IARC) in France, were used as a positive control for PCR reactions. These plasmids were also used in PCR reactions to evaluate the specificity and cross-reactivity of primers for LT-Ag regions of BKPyV and JCPyV. The standards of known copies of plasmids LT-BKPyV-pcDNA3 and LT-JCPyV-pcDNA3 were used as positive controls for the preparation of standard curves of BKPyV and JCPyV, respectively.
Preparation of positive controls for BKPyV and JCPyV
The standard curve of each virus was plotted by using a 10-fold serial dilution of the positive control plasmids i.e. LT-BKPyV-pcDNA3 and LT-JCPyV-pcDNA3 for BKPyV and JCPyV respectively. 10-fold serial dilution of positive control plasmid was prepared according to this formula Copy no of interest = n * NA (n = mass of plasmid required for copy no/molar mass of plasmid, NA = Avogadro’s number).
Real time PCR for the quantification of BKPyV and JCPyV
Real time PCR (qRT-PCR) was performed on PikoReal real time detection system for the detection and absolute quantification of BKPyV as well as JCPyV viral load in sewage samples. qRT-PCR was performed using LT-Ag primers of each virus. The qPCR reaction mixture (10 µl) was prepared using 5 µl of qPCR Master Mix 2x (Thermo Fisher Scientific), 0.25 µM of each forward and reverse primer, 1 µl of 10 mg/ml bovine serum albumin (BSA), 1 µl of the template and molecular biology grade water. The real-time PCR was performed in triplicate for each sample, standard control dilutions (1010 to 102 for BKPyV and 109 to 103 for JCPyV) as well as for negative control. Amplification was performed using initial denaturation at 95 °C for 10 min followed by 40 cycles of denaturation of 30 s at 95 °C, annealing for 30 s at 57 °C, and extension for 30 s at 72 °C with a final extension at 72 °C for 5 min. In qRT-PCR, the melt curve was performed at 55 °C to 95 °C. Melt curve, melt peak and other data analysis were performed with PikoReal2.2 software. The standard amplification curve of the known quantity of plasmid DNA (LT-BKPyV-pcDNA3 and LT-JCPyV-pcDNA3) was used to determine the unknown amount of DNA in the sewage samples.
Results
Samples collection sites
Six main drains of the urban areas of the city of Lahore were selected for the present study based on the declaration as virus hotspots by the local government. These drains were Cantonment drain (at the point of Nishat Colony), Gulshan Ravi drain, Iqbal town drain, Mughalpura drain, Kharak drain (at the point of Sabzazar area), and Roi (or Township) drain. The locations of the sample collection sites are shown in Fig. 1.
Evaluation of primers for BKPyV & JCPyV
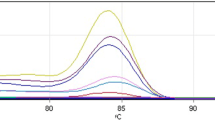
Primer pairs targeting the LT regions of the BKPyV & JCPyV were validated using the plasmids LT-BKPyV-pcDNA3 and LT-JCPyV-pcDNA3, harboring the full-length LT regions of the BKPyV & JCPyV, respectively. Moreover, cross-reactivity of primers for LT-Ag of these viruses was evaluated among BKPyV and JCPyV. The primer set of BKPyV and JCPyV amplified only their targets in respective viruses showing specificity for BKPyV (Fig. 2) and JCPyV (Fig. 3).
Detection of BKPyV and JCPyV in sewage samples
The presence of BKPyV and JCPyV was detected in the DNA extracted from concentrated sewage samples of each drain by performing conventional PCR. The LT-BKPyV-pcDNA3 and LT-JCPyV-pcDNA3 were used as a positive control for BKPyV and JCPyV, respectively. The PCR bands of size 171 bps and 177 bps confirmed the presence of BKPyV (Fig. 4A) and JCPyV (Fig. 4B), respectively in samples while no band appeared in the samples negative for BKPyV and JCPyV. Our data showed that BKPyV and JCPyV were present in all sewage water samples tested at varying concentrations.
PCR based detection of BKPyV (A) and JCPyV (B) in sewage samples targetting LT-Ag region. Lane 1: 1 kb ladder mix; Lane 2: (+) positive control; Lane 3: Iqbal town drain; Lane 4: Gulshan Ravi drain; Lane 5: Kharak drain; Lane 6: Roi drain; Lane 7: Cantonment drain; Lane 8: Mughalpura drain; Lane 9: (-) negative control
Real-time assay for the quantification of BKPyV and JCPyV viral loads
The qPCR amplification of the standards at tenfold serially diluted concentrations of plasmids (LT-BKPyV-pcDNA3 & LT-JCPyV-pcDNA3), was performed to prepare a calibration curve between Ct values and corresponding gene copy numbers for this qPCR assay. The linear range of the qPCR curve corresponded to the Ct values of 2.86 (1010 gene copies) to 27.58 (102 gene copies) and 9.41 (109 gene copies) to 32.64 (103 gene copies) for BKPyV LT-Ag and JCPyV LT-Ag primers, respectively. This curve was used for the quantification of the viral load of BKPyV and JCPyV in the sewage sample (Fig. 5).
Absolute quantification of LT-Ag copies of BKPyV (A) and JCPyV (B) by qPCR in various drains of Lahore. IT (Iqbal Town drain), GR (Gulshan Ravi drain), SZ (Kharak drain), TS (Roi drain), MP (Mughal Pura drain), and NC (Cantonment drain) represent the tested drains of Lahore. Triplicates’ data in the form of dots has been presented here. Different alphabets at samples show the significant differences (p < 0.05) calculated by one way ANOVA and Tukey's multiple comparisons test
Viral loads of BKPyV and JCPyV in sewage samples were measured by comparing their Ct value with standard control dilutions of BKPyV and JCPyV. The viral load as estimated by qPCR targeting LT-Ag regions of the genomes ranged from 1278 to 178368 copies per µg of environmental DNA for BKPyV and 5173 to 79129 copies per µg of environmental DNA for JCPyV.
Discussion
Amongst the known human polyomaviruses, BKPyV and JCPyV are well studied and associated with clinical diseases. BKPyV is linked with nephropathy, ureteral stenosis, and hemorrhagic cystitis whereas progressive multifocal leukoencephalopathy and various neoplasms are associated with JCPyV (Dalianis and Hirsch 2013). Both of these viruses infect humans during early childhood and establish long-lasting asymptomatic latent infection in the immunocompromised hosts. However, intense immunosuppression or immunomodulatory medication in some patients causes reactivation of the virus leading to the cytolytic destruction of the host cells and ultimately release of the virus (Wiedinger et al. 2014). These viruses are mainly detected in the blood, cerebrospinal fluid, and urine of the infected patients. Prolonged excretion of these viruses in urine which is subsequently discharged in the wastewater causes great concern for the environment and could be a source of virus transmission. We planned this study to investigate the presence of BKPyV and JCPyV in the urban sewage of Lahore, Pakistan. Our study being the premier report on the detection of BK and JC polyomaviruses in sewage water described the presence of these viruses in 6 different drains located in various urban areas of the city of Lahore.
Surveillance of wastewater has been proved an efficient way of tracking pathogens including viruses in communities (Hart and Halden 2020). Public health officials not only use this approach to monitor the outbreaks of diseases but also to identify threats to human health. Screening of many viruses including poliovirus, adenovirus, rotavirus, hepatitis A virus, influenza virus, noroviruses, coxsackievirus and coronavirus has been in place in many countries for decades. Very recently, wastewater-based epidemiology (WBE) has been successfully applied for the detection of the spread of SARS-CoV-2 (Farhan Ul Haque et al. 2021; Oliveira et al. 2020). The research on wastewater contamination by human polyomaviruses dates back to 2000, when the detection of BKPyV and JCPyV was first reported in urban sewage of Barcelona (Bofill-Mas et al. 2000). Later on, both of these viruses were detected in urban sewage of various geographical locations including Italy (Iaconelli et al. 2015), Brazil (Comerlato et al. 2016), Australia (Ahmed et al. 2010), and Egypt (Ahmed et al. 2019). In the present study, we reported the detection of BKPyV and JCPyV in sewage water of Lahore for the first time, indicating the ubiquitous presence of BKPyV and JCPyV in the Pakistani population. Our assay detected these viruses in all the sampling points. The possible reasons for this high detection rate could be the dense population and high prevalence of these viruses in the Pakistani population as described previously (Hussain et al. 2017). Moreover, it has been described that the detection rate of BKPyV and JCPyV varies greatly depending on the geographical regions, types of sample, and technique used.
It is worth mentioning here that certain challenges were faced during the execution of this study. The first hurdle was the concentration of viruses as water samples usually contain a low concentration of viruses (Girones et al. 2010). Different methods including skimmed milk flocculation, polyethylene glycol precipitation (PEG), glass wool filtration, ultrafiltration, virus adsorption, and elution have been described for virus concentration (Hoyle 1962; Hjelmsø et al. 2017; Rachmadi et al. 2016). We tried all of these methods for the concentration of the virus and found PEG the most appropriate due to its simplicity, cost-effectiveness, efficiency, and high viral recovery rate. To the best of our knowledge, the PEG method was used for the first time in this study for the concentration of BKPyV and JCPyV in sewage water samples. Extraction of nucleic acids from concentrated sewage water is necessary to detect viruses in sewage water. Various manual and kit-based methods have been reported for the extraction of nucleic acid from environmental samples (Hjelmsø et al. 2017). In this present study, two methods Exgene viral DNA/RNA kit and TRizol method were used for the extraction of nucleic acid. We found that TRizol method is better than Exgene viral DNA/RNA kit as it gave the highest yield of nucleic acids in concentrated sewage water.
Previously culture-based methods were used for the detection of viruses in water, however, these methods could only detect the infectious virus. Moreover, culture-based methods were laborious, time-consuming, and not applicable to all viruses (Haramoto et al. 2018). Recently, various molecular methods have been developed to detect viruses in water including PCR, loop-mediated isothermal amplification (LAMP), nucleic acid sequence-based amplification (NASBA) and enzyme-linked immunosorbent assay (ELISA) (Mattison and Bidawid 2009). Of these methods, PCR is considered the most reliable method for the detection of all types of viruses in water samples (Toze 1999). Initially, in our PCR assay, we targeted the shorter fragments of LT-Ag and VP of both BKPyV and JCPyV. However, LT-Ag based PCR worked efficiently in this study. Furthermore, we used qPCR for the quantification of the BKPyV and JCPyV in the six drains samples. Our qPCR displays high sensitivity with detection limits 102 and 103 gene copy numbers for BKPyV and JCPyV, respectively.
Our data showed an overall high viral load of JCPyV in comparison to BKPyV in the samples from all drains except for Gulshan Ravi drain (GR) sample. This observation suggests the high prevalence and active infection of JCPyV in the Pakistani population in most areas. Interestingly, the shedding of JCPyV was described at a higher rate when compared with BKPyV, in the urine samples of healthy individuals in different geographical locations of the world (Boukoum et al. 2016; Urbano et al. 2016). Bofill-Mas et al. (2000) also reported that the concentration of JCPyV is higher than BKPyV in sewage water. The higher concentration of JCPyV is due to more excretion through urine not only in immunocompromised individuals but also in immunocompetent individuals (Rachmadi et al. 2016). Moreover, Fumian et al. (2010) reported that the higher prevalence of JCPyV in sewage water is due to its higher stability in the environment. The highest viral load of JCPyV was observed in the Kharak drain (SZ) while for BKPyV the highest load was observed in Gulshan Ravi (GR) drain. Overall, the viral load in this study ranged from 1278 to 178368 copies per µg of environmental DNA for BKPyV and 5173 to 79129 copies per µg of environmental DNA for JCPyV. These results are in line with previous studies where high viral loads of JCPyV and BKPyV were described (Albinana-Gimenez et al. 2009; McQuaig et al. 2009). The difference in viral loads among these drains can be explained by the differences in population density in the surrounding regions of the drains.
Here, we reported the detection of BKPyV and JCPyV in sewage water collected from various drains in urban areas of Lahore, Pakistan showing the high prevalence of JCPyV and BKPyV in the Pakistani population. We speculate that detection of BKPyV and JCPyV in sewage water may offer a good strategy to measure the true scale of these viruses' epidemiology in the Pakistani population. However, further studies are required to develop a better understanding of the epidemiology and pathobiology of these viruses in Pakistan.
References
Ahmed W, Wan C, Goonetilleke A, Gardner T (2010) Evaluating sewage-associated JCV and BKV polyomaviruses for sourcing human fecal pollution in a coastal river in Southeast Queensland, Australia. J Environ Qual 39:1743–1750. https://doi.org/10.2134/jeq2010.0062
Ahmed NI, Elmahdy EM, Allayh AK, Mohamed El-CB, Loutfy SA, Barakat A, Ali MA (2019) Prevalence of human polyomavirus and papillomavirus in wastewater and in stool of Egyptian patients. Egypt J Aquatic Biol Fish 23:29–41. https://doi.org/10.21608/ejabf.2019.29237
Albinana-Gimenez N, Miagostovich MP, Calgua B, Huguet JM, Matia L, Girones R (2009) Analysis of adenoviruses and polyomaviruses quantified by qPCR as indicators of water quality in source and drinking-water treatment plants. Water Res 43:2011–2019. https://doi.org/10.1016/j.watres.2009.01.025
Barbanti-Brodano G, Martini F, De Mattei M, Lazzarin L, Corallini A, Tognon M (1998) BK and JC human polyomaviruses and simian virus 40: natural history of infection in humans, experimental oncogenicity, and association with human tumors. Adv Virus Res 50:69–99. https://doi.org/10.1016/S0065-3527(08)60806-4
Bofill-Mas S, Pina S, Girones R (2000) Documenting the epidemiologic patterns of polyomaviruses in human populations by studying their presence in urban sewage. Appl Environ Microbiol 66:238–245. https://doi.org/10.1128/AEM.66.1.238-245.2000
Boukoum H, Nahdi I, Sahtout W, Skiri H, Segondy M, Aouni M (2016) BK and JC virus infections in healthy patients compared to kidney transplant recipients in Tunisia. Microb Pathog 97:204–208. https://doi.org/10.1016/j.micpath.2016.06.015
Bouvard V, Baan RA, Grosse Y, Lauby-Secretan B, El-Ghissassi F, Benbrahim-Tallaa L, Guha N, Straif K (2012) Carcinogenicity of malaria and of some polyomaviruses. Lancet Oncol 13:339–340. https://doi.org/10.1016/S1470-2045(12)70125-0
Ciotti M, Prezioso C, Pietropaolo V (2019) An overview on human polyomaviruses biology and related diseases. Futur Virol 14:487–501. https://doi.org/10.2217/fvl-2019-0050
Comerlato J, Souza-Campos F, Souza-Arantes T, Roos-Kulmann M, Trindade-Oliveira M, Rosado-Spilki F, Guedes-Frazzon A, Roehe PM, Franco AC (2016) Distribution and genetic diversity of the human polyomaviruses JC and BK in surface water and sewage treatment plant during 2009 in Porto Alegre, Southern Brazil. Braz J Biol 77:459–468. https://doi.org/10.1590/1519-6984.15415
Condez AC, Nunes M, Filipa-Silva A, Leonardo I, Parreira R (2021) Human Polyomaviruses (HPyV) in Wastewater and Environmental Samples from the Lisbon Metropolitan Area: Detection and Genetic Characterization of Viral Structural Protein-Coding Sequences. Pathogens 10:1309. https://doi.org/10.3390/pathogens10101309
Dalianis T, Hirsch HH (2013) Human polyomaviruses in disease and cancer. Virology 437:63–72. https://doi.org/10.1016/j.virol.2012.12.015
De Gascun CF, Carr MJ (2013) Human polyomavirus reactivation: disease pathogenesis and treatment approaches. Clin Dev Immunol 2013:1–27. https://doi.org/10.1155/2013/373579
Dehcheshmeh LK, Makvandi M, Timori A (2020) Prevalence of human polyomavirus JC and BK in normal population. Asian Pac J Cancer Prev 21:2877. https://doi.org/10.31557/APJCP.2020.21.10.2877
Egli A, Infanti L, Dumoulin A, Buser A, Samaridis J, Stebler C, Gosert R, Hirsch HH (2009) Prevalence of polyomavirus BK and JC infection and replication in 400 healthy blood donors. J Infect Dis 199:837–846. https://doi.org/10.1086/597126
Farhan Ul Haque M, Bukhari SS, Ejaz R, Zaman FU, Sreejith KR, Rashid N, Umer M, Shahzad N (2021) A novel RdRp-based colorimetric RT-LAMP assay for rapid and sensitive detection of SARS-CoV-2 in clinical and sewage samples from Pakistan. Virus Res 302:198484. https://doi.org/10.1016/j.virusres.2021.198484
Fumian TM, Guimarães FR, Pereira-Vaz BJ, Da-Silva MTT, Muylaert FF, Bofill-Mas S, Gironés R, Leite JPG, Miagostovich MP (2010) Molecular detection, quantification and characterization of human polyomavirus JC from waste water in Rio De Janeiro, Brazil. J Water Health 8:438–445. https://doi.org/10.2166/wh.2010.090
Girones R, Ferrus MA, Alonso JL, Rodriguez-Manzano J, Calgua B, Correa ADA, Hundesa A, Carratala A, Bofill-Mas S (2010) Molecular detection of pathogens in water–the pros and cons of molecular techniques. Water Res 44:4325–4339. https://doi.org/10.1016/j.watres.2010.06.030
Haramoto E, Kitajima M, Hata A, Torrey JR, Masago Y, Sano D, Katayama H (2018) A review on recent progress in the detection methods and prevalence of human enteric viruses in water. Water Res 135:168–186. https://doi.org/10.1016/j.watres.2018.02.004
Hart OE, Halden RU (2020) Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: Feasibility, economy, opportunities and challenges. Sci Total Environ 730:138875. https://doi.org/10.1016/j.scitotenv.2020.138875
Hjelmsø MH, Hellmér M, Fernandez-Cassi X, Timoneda N, Lukjancenko O, Seidel M, Elsässer D, Aarestrup FM, Löfström C, Bofill-Mas S (2017) Evaluation of methods for the concentration and extraction of viruses from sewage in the context of metagenomic sequencing. PLoS One 12:e0170199. https://doi.org/10.1371/journal.pone.0170199
Hoyle L (1962) Book reviews: partition of cell particles and macromolecules: distribution and fractionation of cells, viruses, microsomes, proteins, nucleic acids, and antigen-antibody complexes in aqueous polymer two-phase systems, by Per-Ake Albertsson, p 231. ALMQVIST & WIKSELL, Stockholm (JOHN WILEY & SONS, New York & London), 1961. £2 16s. 0d. [IQ/71]. R Soc Health J 82(1):31–31. https://doi.org/10.1177/146642406208200129
Hussain I, Tasneem F, Umer M, Pervaiz A, Raza M, Arshad MI, Shahzad N (2017) Specific and quantitative detection of Human polyomaviruses BKPyV and JCPyV in the healthy Pakistani population. Virol J 14:1–11. https://doi.org/10.1186/s12985-017-0752-2
Iaconelli M, Petricca S, Libera SD, Di Bonito P, La Rosa G (2015) First detection of human papillomaviruses and human polyomaviruses in river waters in Italy. Food Environ Virol 7:309–315. https://doi.org/10.1007/s12560-015-9203-7
Kamminga S, van der Meijden E, Feltkamp MC, Zaaijer HL (2018) Seroprevalence of fourteen human polyomaviruses determined in blood donors. PLoS One 13:e0206273. https://doi.org/10.1371/journal.pone.0206273
Kean JM, Rao S, Wang M, Garcea RL (2009) Seroepidemiology of Human Polyomaviruses. PLoS Pathog 5:1000363. https://doi.org/10.1371/journal.ppat.1000363
Khalili K, Sariyer IK, Safak M (2008) Small tumor antigen of polyomaviruses: role in viral life cycle and cell transformation. J Cell Physiol 215:309–319. https://doi.org/10.1002/jcp.21326
Limaye AP, Jerome KR, Kuhr CS, Ferrenberg J, Huang ML, Davis CL, Corey L, Marsh CL (2001) Quantitation of BK virus load in serum for the diagnosis of BK virus–associated nephropathy in renal transplant recipients. J Infect Dis 183:1669–1672. https://doi.org/10.1086/320711
Linthorst J, Welkers MR, Sistermans EA (2022) Clinically relevant DNA viruses in pregnancy. Prenat Diagn 2022:1–10. https://doi.org/10.1002/pd.6116
Mattison K, Bidawid S (2009) Analytical methods for food and environmental viruses. Food Environ Virol 1:107–122. https://doi.org/10.1007/s12560-009-9017-6
McQuaig SM, Scott TM, Lukasik JO, Paul JH, Harwood VJ (2009) Quantification of human polyomaviruses JC virus and BK virus by TaqMan quantitative PCR and comparison to other water quality indicators in water and fecal samples. Appl Environ Microbiol 75:3379–3388. https://doi.org/10.1128/AEM.02302-08
Nomburg J, Zou W, Frost TC, Datta C, Vasudevan S, Starrett GJ, Imperiale MJ, Meyerson M, DeCaprio JA (2022) Long-read sequencing reveals complex patterns of wraparound transcription in polyomaviruses. PLoS Pathog 18:e1010401. https://doi.org/10.1371/journal.ppat.1010401
Oliveira MDLA, Campos A, Matos AR, Rigotto C, Sotero-Martins A, Teixeira PF, Siqueira MM (2020) Wastewater-based epidemiology (WBE) and viral detection in polluted surface water: A valuable tool for COVID-19 surveillance—A brief review. Int J Environ Res Public Health 17:9251. https://doi.org/10.3390/ijerph172492511
Rachmadi AT, Torrey JR, Kitajima M (2016) Human polyomavirus: Advantages and limitations as a human-specific viral marker in aquatic environments. Water Res 105:456–469. https://doi.org/10.1016/j.watres.2016.09.010
Šroller V, Hamšíková E, Ludvíková V, Musil J, Němečková Š, Saláková, M (2016) Seroprevalence rates of HPyV6, HPyV7, TSPyV, HPyV9, MWPyV and KIPyV polyomaviruses among the healthy blood donors. J Med Virol 88:1254–1261. https://doi.org/10.1002/jmv.24440
Toze S (1999) PCR and the detection of microbial pathogens in water and wastewater. Water Res 33:3545–3556. https://doi.org/10.1016/S0043-1354(99)00071-8
Urbano PRP, Oliveira RR, Romano CM, Pannuti CS, Fink MCDDS (2016) Occurrence, genotypic characterization, and patterns of shedding of human polyomavirus JCPyV and BKPyV in urine samples of healthy individuals in São Paulo, Brazil. J Med Virol 88:153–158. https://doi.org/10.1002/jmv.24318
Van Ghelue M, Khan MTH, Ehlers B, Moens U (2012) Genome analysis of the new human polyomaviruses. Rev Med Virol 22:354–377. https://doi.org/10.1002/rmv.1711
Whiley DM, Mackay IM, Sloots TP (2001) Detection and differentiation of human polyomaviruses JC and BK by LightCycler PCR. J Clin Microbiol 39:4357–4361. https://doi.org/10.1128/JCM.39.12.4357-4361.2001
Wiedinger K, Bitsaktsis C, Chang S (2014) Reactivation of human polyomaviruses in immunocompromised states. J Neurovirol 20:1–8. https://doi.org/10.1007/s13365-014-0234-x
Acknowledgements
This work was supported by the research project grant from the University of the Punjab, Lahore.
Author information
Authors and Affiliations
Contributions
Conceptualization: [Muhammad Farha Ul Haque and Naveed Shahzad]
Methodology: [Rabia Ejaz, Muhammad Farha Ul Haque and Naveed Shahzad]
Formal analysis and investigation: [Rabia Ejaz, Muhammad Farha Ul Haque and Naveed Shahzad],
Writing—original draft preparation: [Rabia Ejaz, Muhammad Farha Ul Haque and Naveed Shahzad]
Writing—review and editing: [Muhammad Farha Ul Haque and Naveed Shahzad]
Resources: [Muhammad Farha Ul Haque and Naveed Shahzad]
Supervision: [Muhammad Farha Ul Haque and Naveed Shahzad]
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ijaz, R., Shahzad, N. & Farhan Ul Haque, M. Detection of BK and JC polyomaviruses in sewage water of the urban areas of Lahore, Pakistan. Biologia 78, 2895–2902 (2023). https://doi.org/10.1007/s11756-023-01430-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11756-023-01430-6