Abstract
Edwardsiella tarda is one of the serious threats affecting the worldwide aquaculture. In the present study, four isolates were recovered from diseased goldfish, showing hemorrhages, reported with 60% mass mortality in an ornamental fish farm, Ernakulam, Kerala. Based on the phenotypic and genotypic analysis, the bacteria were identified as Edwardsiella tarda, Citrobacter freundii, Acinetobacter junii and Comammonas testosteronii. Experimental challenge studies using healthy goldfish revealed that among the four isolates, E. tarda alone leads to 100% mortality of experimental fish within 175 degree days and the pathogen could be successfully re-isolated from the moribund fish. The LD50 value of E. tarda was calculated as 9.9 × 105 CFU/fish. The histopathology of the infected tissues of goldfish had shown the typical features of E .tarda infection. The pathogen was found positive for the virulence genes viz., hly, etfA, etfD and eseD as detected using PCR. Thus E. tarda was confirmed as the real causative agent of the disease outbreak. Multiple antimicrobial resistance (AMR) exhibited by the pathogen towards 19 tested antibiotics with the MAR index of 0.46 highlighted the exposure of antibiotics to the fish in the farm. The existence of antibiotic resistant genes within the plasmid as revealed through plasmid curing studies pointed out the possibility of rapid dissemination of AMR in aquaculture. Hence proper surveillance and appropriate diagnostic methods need to be implemented at regular intervals to mitigate the menace.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ornamental fish industry faces diverse bacterial diseases which affect the successful farming and lead to huge economic losses worldwide. Goldfish (Carassius auratus) is one of the most important domesticated ornamental fish in aquaria and are recently reported to be infected with various bacterial species like Aeromonas, Acinetobacter, Edwardsiella, Lactococcus etc. (Preena et al. 2019) thereby interrupting the sustainable fish culture. Mass mortality of goldfish population infected with Aeromonas hydrophila (Dharmaratnam et al. 2018), A. salmonicida (Jin et al. 2020), Proteus hauseri (Kumar et al. 2015), Mycobacterium marinum, (Hodgkinson et al. 2012), Flavobacterium columnare (Verma et al. 2015) etc. was noticed in various fish farms. Multidrug resistant enterobacteriaceae microbial groups such as Edwardsiella tarda, E.coli, Kluyvera cryocrescens, Enterobacter cloacae, Plesiomonas shigelloides, Citrobacter freundii, Klebsiella aerogenes, Providencia vermicola Proteus hauseri, Proteus pinneri and Serratia marcescens were recently reported to induce virulence thereby causing severe infection in goldfish (Preena et al. 2020b). Amidst these pathogens, Edwardsiella is one of the major threats in aquaculture, which comprises mainly five species; E. tarda, E. ictaluri, E. hoshinae, E. piscicida and E. anguillarum (Austin and Austin 2016; Kerie et al. 2019). Out of which, Edwardsiella tarda is a well-known pathogen and etiological agent for major diseases in the fresh and brackish water aquaculture system (Shetty et al. 2014). Davies et al. (2018) reported that E. tarda is one of the major causes of severe disease outbreaks in fish farming industry and also poses high zoonotic risks. It has been initially characterized by Trabulsi and Ewing (1962). It was reported to induce severe diseases viz., hemorrhagic septicaemia, skin lesions and can also affect internal organs thereby leading to higher mortalities in various fish species (Dubey et al. 2018). Edwardsiellosis is a major putrefactive disease caused by E. tarda which affects many fishes like tilapia, catfish, salmon, trout etc. (Mohanty and Sahoo 2007). However, the reports of E. tarda as significant pathogen in ornamental fish are very few (Walczak et al. 2017). Being zoonotic, E. tarda raise the chance of dissemination to humans thereby posing health threats (Leung et al. 2012). Hence timely detection of the infection through proper surveillance is essential in aquaculture systems.
The broad host range, wide distribution and presence of significant virulence factors enable Edwardsiella to induce severe pathogenesis in diverse hosts (Park et al. 2012). Several motility-related proteins such as fimbrial adhesin-like protein play an important role in inducing virulence by penetrating into the host epithelial cells (Sakai et al. 2009). Besides, certain extracellular hemolysin proteins are also crucial in invading host cells thereby promoting pathogenesis (Wang et al. 2010a). The emergence of antimicrobial resistance (AMR) in E. tarda enhances the virulence and elevates the risk in successful treatment of the disease (Yu et al. 2012). The existence of antibiotic resistant genes; whether plasmid or chromosome, can be located using plasmid curing studies (Preena et al. 2020b). The present study was focused to investigate the cause of disease outbreak in a goldfish farm. Further, the isolated pathogen was subjected to phenotypic and genotypic characterization for determining their virulence and antimicrobial resistance and the infectivity was confirmed through in-vivo study.
Materials and methods
Fish sampling and screening of bacterial pathogens
Ornamental goldfish (Carassius auratus) (n=5) showing hemorrhagic septicemia and gill paleness were collected from a farm, Ernakulam, Kerala with 60% cumulative mortality. The geographic map showing the sampling site of infected goldfish is given in Fig. 1. Under aseptic conditions, the infected tissues such as gills, kidney, liver and spleen were excised separately from the five diseased fish. Then the tissues were homogenized and inoculated into nutrient broth (HiMedia) and incubated for 24 h at 28 °C for bacterial isolation. The overnight grown cultures were serially diluted and spread plated on trypticase soy agar (HiMedia). The unique isolated colonies were subjected to biochemical tests viz., Gram’s staining, motility, catalase, oxidase, urease, H2S production, nitrate reduction, Indole production, Methyl red, Voges–Proskauer, Simmon’s Citrate, lysine decaboxylase, sugar fermentation from glucose, lactose and sucrose according to Barrow and Feltham (2004). The cultures were also inoculated on MacConkey agar plates to observe their fermentation property. The test results for suspected E. tarda were compared with the E. tarda type strain (ATCC 15,947T) and reference strain as defined by Ewing et al. (1965). After preliminary identification, the isolates were stored at −80 °C in 30% (v/v) glycerol for further phenotypic and genotypic characterization.
DNA extraction, PCR amplification and phylogenetic analyses
Genomic DNA of the unique bacterial isolates was extracted using salting out method (Miller et al. 1988). The quantity and quality of the extracted DNA was determined using UV spectrophotometer (Beckman, USA) at the absorbance of 260nm and 280nm. Bacterial 16srRNA universal primers fD1 (5’CCG AAT TCG TCG ACA ACA GAG TTT GAT CCT GGC TCA G3’) and rD1 (5’CCC GGG ATC CAA GCT TAA GGA GGT GAT CCA GCC3’) were employed for PCR amplification under the conditions described by Weisburg et al. (1991). The 16srRNA gene PCR products were sequenced using Sanger’s method at Scigenom Pvt., Ltd., India. The obtained sequences of four bacterial strains were compared using NCBI BLAST (Basic Local Alignment Search Tool) and RDP (Ribosomal Database Project).
Experimental challenge studies
Fifty healthy goldfish were purchased from an ornamental fish aquarium, Ernakulam. They were acclimatized for 10 days in a 500 L capacity glass aquaria containing 400 L water. The temperature was maintained at 28 °C, pH at 7.2-8.1 and continued aeration was provided with daily exchange of 50% water. All the four bacterial isolates were revived from glycerol stocks stored at -80 °C and inoculated on TSA plates and incubated overnight at 28 °C. Bacterial suspensions were prepared by further inoculating the trypticase soy broth with single pure colony of each isolate and kept under shaker incubator for overnight at 28 °C. The experimental and control groups contained 10 fish each and they were injected intra muscularly with the four bacterial suspensions such as E. tarda (NPET-1), Citrobacter freundii (NPCF-1), Acinetobacter junii (NPAJ-1) and Comamonas testosteroni (NPCT-1) at the rate of 2.3 × 107, 2.7 × 107, 2.6 × 107 and 3.1 × 107 CFU/ml respectively. While the ten fish of control groups were injected with sterile normal saline. After injection, each group was kept under observation for two weeks; the mortality and survival rate were monitored daily. The affected tissues were taken for bacterial re-isolation and phenotypic and genotypic tests were repeated to confirm the primary pathogen.
The 50% lethal dose (LD50) of E. tarda was calculated according to the method described by Reed and Muench (1938). The ten-fold serially diluted bacterial cell suspension (0.1ml) in sterile saline at the range of 104 to 107 CFU/ml was intra muscularly injected into experimental fish. Sterile saline (0.1ml) treated fish samples were kept as control. The treated fish were monitored daily for checking the morbidity and mortality.
Histological analysis
The infected internal organs such as kidney, spleen and liver of the experimentally challenged fish were excised and preserved in 10% neutral buffered formalin for histopathological analysis. The samples were fixed in paraffin wax and stained with haematoxylin and eosin (H&E). The analysis was performed following Chinabut et al. (1995).
Screening of virulence genes and tarda specific gene
The isolate which has been identified as E. tarda (NPET-1) was screened for the presence of virulence genes such as etfA and etfD which encode for hemagglutinating fimbrial major subunits, hly (hemolysin), ent hly (enterohemolysin), and TTSS (Type III Secretion System) genes viz., eseB, eseC, eseD, esaC, escA, esrA and esrB using PCR. The gene specific for E. tarda namely tarda was also checked for species level confirmation. The PCR reaction was performed with the total reaction volume of 20 µl, having 10 µl of master mix (Takara), 1 µl of DNA template, 0.5 µl of forward and reverse primers specific for each gene and 8 µl of sterile MilliQ in PCR thermal cycler (Eppendorf, India). The details showing the primers and PCR amplification conditions are listed in Table 1.
Antibiogram profiling before and after plasmid curing
Antimicrobial susceptibility tests were performed for the NPET-1 isolate using disc diffusion on Mueller–Hinton agar (HiMedia) with 46 antibiotics of 15 classes as listed in Table 2. The resistance and susceptibility of the isolate were recorded by measuring the diameter of the inhibition zone (mm) as per the guidelines of Clinical and Laboratory Standards Institute (CLSI 2020). The minimum inhibitory concentration (MIC) of antibiotics was also determined using MIC strips embedded with a different series of antibiotic concentration. A CLSI reference strain, E. coli ATCC 25,922 was kept as the positive control and 0.5 McFarland solution as the standard for getting accurate results.
The plasmid of the NPET-1 isolate was extracted using plasmid mini preparation kit (Sigma Aldrich, USA). After assuring the presence of plasmid by running the eluted plasmid on 1% agarose gel, plasmid curing has been done for the isolate using acridine orange (AO) (Sigma Aldrich, USA). For this, a different concentration of AO (0.1-1 mg/ml) was added to nutrient broth inoculated with the isolate and kept incubated overnight at 28 °C. After plasmid curing, plasmid extraction was repeated to confirm the elimination of plasmid. Further, the antibiotic susceptibility tests were again carried out to observe the change in antibiogram profile after plasmid curing so as to determine the location of antibiotic resistance gene (ARG).
Results and discussion
Bacterial isolation and identification
Several colonies were isolated from the tissues such as gills, liver, kidney and spleen of all the infected fish. Based on the biochemical characters, four unique isolates were identified and the taxonomic status of the bacteria was revealed through 16SrRNA gene sequence analysis. The isolates showed 100% homology to E. tarda (NPET-1), C. freundi (NPCF-1), A. junii (NPAJ-1) and C. testosteroni (NPCT-1). The most dominant isolate recovered from all the tissues of five infected fish were E. tarda and which appeared as small, circular, convex transparent colonies of around 0.4mm in diameter on TSA plates. While, C. freundi was segregated from liver and spleen, C. testosteroni from gills, liver and kidney and A .junii from gills, kidney and spleen. All the cultures had grown on MacConkey agar plates as white non-lactose fermenting colonies except C.freundi which appeared as pink lactose fermenting colonies. Among the 15 biochemical indexes, the NPET-1 isolate was tested as Gram negative, short rods, motile and was positive to catalase, indole production, methyl red, H2S production, glucose fermentation, nitrate reduction and lysine decarboxylase and negative to all other tests. The biochemical test results of E.tarda (NPET-1) were in close accordance with the type strain ATCC 15,947T and the reference strain (Ewing et al. 1965). The co-infection of goldfish with E.tarda, C. freundii, Acinetobacter and Comamonas in causing pathogenicity were recently reported by Preena et al. (2019). Previously, C. freundii was reported as one of the major causative agents of clinical illness in cyprinids (Jeremi et al. 2003). The co-occurrence of E. tarda, C. freundii and A.junii in diseased freshwater ornamental fish was noticed by Walczak et al. (2017) also. Since all the identified isolates were reported as pathogens in ornamental fish industry, identifying the real causative agent is the utmost necessity.
Phylogenetic analysis of the isolates
The 16srRNA gene sequences with 1450 bp size of NPET-1, NPAJ-1, NPCT-1 and NPCF-1 were determined by alignment using BLAST and RDP and were submitted to Genbank with accession numbers: MZ467314, MZ467315, MZ467316 and MZ467317 respectively. The phylogenetic tree of the pathogenic isolate E.tarda (NPET-1) was constructed (Fig. 2) with the distance coefficient of 0.004 to 0.000 using sequences of E. tarda and that of other species such as E. ictaluri, E. piscicida, E. anguillarum and E. hoshinae retrieved from RDP database. The sequence alignments and phylogeny were studied using Mega X software (Kumar et al. 2018). The tree revealed that NPET-1 exhibited high sequence homology to E. tarda and were clustered together. The pairwise distance between NPET-1 and E. tarda determined using Kimura2 parameter was 0.00132. While the distance value between NPET-1 and other species of same genus such as E. ictaluri, E. piscicida, E. anguillarum and E. hoshinae were 0.00733, 0.0068572620, 0.0074889177 and 0.0013655377 respectively. The tree showed that E. tarda NPET-1 showed sequence similarity to E. hoshinae also while compared to other species.
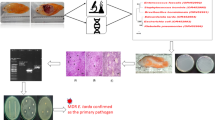
Experimental challenge trials and histological changes in moribund goldfish
The experimentally challenged goldfish exhibited clinical symptoms after 4 days like gill paleness, spleen enlargement, hemorrhage and septicemia throughout the body and in internal organs as reported in naturally infected goldfish obtained from the farms (Fig. 3). A 100% cumulative mortality was observed in the goldfish injected with E. tarda within 175 degree days while 100% survival was noticed in those experimentally challenged with C.freundii, A.junii and C.testosteroni and control groups up to 350 degree days post challenge. Thus it has been concluded that E. tarda was the causative agent and responsible for the disease outbreak. The infected pathogen, E. tarda was successfully isolated from the affected tissues of the dead fish thereby satisfied the Koch’s postulates. The recovered isolates displayed homologous phenotypic and genotypic characteristics of the original isolate, NPET-1. The LD50 value was calculated as 9.9 × 105 CFU/fish by observing the cumulative mortality rate of 30%, 50%, 70% and 100% in goldfish challenged with 104, 105, 106 and 107 CFU respectively of NPET-1. E. tarda was implicated in various disease outbreaks and reported as one of the serious bacterial pathogens in global aquaculture industry (Reichley et al. 2017). Challenge studies have been employed in the earlier studies to determine the virulence of the bacterial pathogens like Aeromonas veronii in Nile tilapia (Raj et al. 2019). Significant histopathological changes were observed in liver, kidney and spleen of the moribund fish experimentally challenged with NPET-1 (Fig. 4). Section of kidney showed diffused interstitial nephritis with tubular degeneration and multifocal areas of necrosis with karyolysis. Suppurative interstitial nephritis is the particular histopathological characteristics of edwardsiellosis in fishes (Miyazaki and Egusa 1976). Besides, ischemic necrosis of tubular epithelium and rupture of basement membrane was also observed. Section of liver exhibited multifocal necrotic hepatitis with diffused vacuolar degeneration of hepatocytes and diffused sinusoidal dilatation and congestion. Lymphocytolysis was noticed in spleen with melanomacrophage aggregation. Interstitial nephritis, necrotic hepatitis and increased melanomacrophage reaction are the typical histopathological features observed in E. tarda infected tissues in previous analyses (Mohanty and Sahoo 2007), which confirmed the severe infection of E.tarda in the experimentally challenged goldfish.
Histopathological changes in (A) Kidney: Thick arrow: tubular degeneration & necrosis; Arrowhead: interstitial nephritis; Thin arrow: rupture of basement membrane (B) Liver: Thick arrow: multifocal areas of necrosis of the hepatic parenchyma; Thin arrow: vacuolar degeneration; Star: Increased activity of kupffer cells; arrowhead: sinusoidal dilatation (C) Spleen: Arrow: Multifocal areas of lymphocytolysis of the spleen
Detection of virulence and species specific genes
In addition to 16srRNA sequence analysis, the studied NPET-isolate was confirmed as E. tarda species through the positive amplification of tarda gene with the product size of 1109 bp. Upon screening of eleven virulence genes using PCR, four were found to be positive for the pathogen. Hemolysin (hly) gene, both fimbrial protein encoding genes (etfA and etfD) and TTSS effector protein encoding gene (eseD) were successfully amplified with the product size of 1177 bp, 415 bp, 445 bp and 391 bp respectively (Fig. 5). Even though hemolysin was responsible for the virulence in the NPET-1, enterohemolysin was not encompassed within them. Out of the various virulence factors of E. tarda, hemolysin is the major protein associated with the invasion and cytotoxicity of the pathogen (Verjan et al. 2005). Shen and Chen (2005) stated that the expression of hly gene in E.tarda lead to the increased mortality of infected fish. Hence the presence of hly gene in the studied isolate pointed out the significance of their virulence and thereby inducing severe pathogenicity in the host. Among the seven type III secretion system genes tested, only one gene (eseD) was found to be present in NPET-1 E. tarda. Type III secretion system play a vital role in inducing pathogenesis for E. tarda by releasing the virulence factors into the host for subverting the normal cell functions. However in this study genes encoding TTSS regulator (esrA and esrB), apparatus (esaC) and chaperon proteins (escA) were absent from the isolate and only those encoding TTSS effector (eseD) was amplified. Without the presence of any one of TTSS virulence factors, E.tarda failed to induce pathogenicity in the host (Tan et al. 2005). It was also demonstrated that eseD mutant pathogen exhibited reduced proliferation and tenfold decrease in virulence in fishes (Wang et al. 2010b). Because of the significance of eseD in inducing pathogenicity, eseD protein acts as the candidate antigen for vaccine development against edwardsiellosis (Wang et al. 2010b). The presence of both fimbrial major subunit genes (etfA and etfD) indicated the enhanced hemagglutinating activity of the studied strain (Sakai et al. 2003). The adhesins located on the fimbrial tip of the pathogen mediates the key step in pathogenicity like adherence to host (Sakai et al. 2004). Decreased virulence was noticed in fishes infected with etfA mutant E. tarda (Sakai et al. 2007). The presence of all identified virulence genes highlighted the infectivity of NPET-1 isolate to the goldfish.
Antibiotic resistance pattern before and after plasmid curing
Amidst the fifteen classes tested, E.tarda NPET-1 exhibited resistance towards nineteen antibiotics of ten different classes. The isolate had shown resistance towards at least one antibiotic belonging to beta lactams, first, second and third generation cephalosporins, aminoglycosides, phenicols, tetracyclines, macrolides, glycopeptides and other classes. The emergence of resistance towards the new generation cephalosporins raises the major concern in disease treatment. Meanwhile, the pathogen had shown no resistance towards any of the tested antibiotics of fourth generation cephalosporins, carbapenems, quinolones and nitrofuran groups. The minimum inhibitory concentration of most of the resistant antibiotics was found to be >256mcg/µL. The multiple antibiotic resistance (MAR) index for the isolate was calculated as 0.46. A MAR index of > 0.2 indicated the heavy exposure of antibiotics to the fish in the farm (Krumperman 1983). Thus the elevated MAR index denotes the excess entry of antimicrobial residues to the farm under investigation. It was reported that E.tarda is naturally resistant to macrolides and glycopeptides (Stock and Wiedemann 2001) which is in close agreement with the present finding. Niu et al. (2019) also documented the multidrug resistance of E. tarda associated with Tilapia towards tetracyclines, sulphonamides and beta-lactams; which supported the present finding. The establishment of multiple antimicrobial resistance in the fish pathogens affects the successful antibiotic treatment.
Unraveling the exact location of AMR genes in the pathogenic bacteria is essential to determine the extent of AMR spread. Most of the antibiotic resistant genes, transposons, integrons and potential virulence genes are encoded within the mobile genetic elements (MGE) such as plasmid (Preena et al. 2020a). After confirming the presence of plasmid in the isolate after plasmid extraction, plasmid curing has been done. After assuring the plasmid elimination, antibiotic susceptibility tests were repeated to determine whether the AMR genes were plasmid borne or chromosome borne. A drastic change in the antibiogram profile was observed after plasmid curing. The isolate had shown resistance towards only three antibiotics such as ampicillin, amoxicillin and colistin. Accordingly the MAR index of the isolate was largely reduced to 0.073. This pointed out that majority of the antibiotic resistant genes existed within the MGE of the pathogen and hence increases the chance of AMR dissemination through horizontal gene transfer (HGT) among the cultured fishes (Preena et al. 2020a). Similar to the present study, a multidrug resistant E. tarda was recovered from the diseased goldfish with the MAR index of 0.35 and after plasmid curing, MAR index was severely reduced to 0.04 (Preena et al. 2020b). In a previous study, a major finding observed was that virulence to fish has been mainly contributed by the large antibiotic-resistance plasmid integrated within E.tarda (Yu et al. 2012). Thus the identification of antibiotic resistant genes carrying plasmid within the pathogen corroborated the increased virulence of NPET-1. The existence of antimicrobial resistant genes within the mobile genetic element of the virulent pathogen, E. tarda elevates the risk of transfer of AMR genes to the whole ecosystem. This may undoubtedly raise threats to human also as E. tarda could induce severe infections in wide range of hosts including human beings (Xu and Zhang 2014).
It was reported in a very earlier study that E. tarda introduced serious exotic diseases in Australian fighting fish thereby spreading the pathogen worldwide (Humphrey et al. 1986). Apart from a very few reports of identification of E. tarda among the diseased ornamental fishes viz. goldfish (Preena et al. 2020b) and koi carp (Preena et al. 2019), disease outbreaks of E. tarda in pet fishes are hardly noticed. Hence the current study provides valuable information regarding the pathogenicity of multi drug resistant E. tarda in ornamental fish industry. However, E.tarda was reported to an intriguing problem in aquaculture worldwide by causing edwardsiellosis in cultured fishes such as tilapia, silver carp, catfish, turbot, Japanese flounder, European eel etc. (Xu and Zhang 2014). This identified pathogen is not only restricted to the cultured fishes but also noticed among the wild fishes like European eel in natural wetland environment (Alcaide et al. 2006). All of this information highlighted the global occurrence of E. tarda and their rapid dissemination among the aquatic environment and therefore their occurrence in ornamental fish farm is also not surprising. Very recently, in accordance with the present study, E. tarda from infected Nile Tilapia and African Catfish of Egyptian fish farm exhibited multidrug resistance towards six antimicrobial classes and found to harbor potential virulence and AMR determinants (Algammal et al. 2021). Thus the existence of multi drug resistant pathogens throughout the world raises major concern in successful treatment of bacterial diseases in aquaculture industry. Therefore implementation of proper monitoring programs, effective precautions and alternative strategies to overcome the menace in aquaculture is the need of the hour.
Conclusions
Pathogenic Edwardsiella tarda harboring significant virulence genes along with the secondary pathogens viz., Acinetobacter juni, Citrobacter freundii and Comammonas testosteroni were recovered from the diseased goldfish. E.tarda was confirmed as the real causative agent as it causes 100% mortality when experimentally challenged and it should be considered as the potential threat in goldfish farms. Unique histopathological features of E.tarda infection were noticed in the infected tissues of the experimentally challenged goldfish. Identification of potential virulence and plasmid borne multi drug resistance in the studied pathogen challenges the effective antibiotic treatment and promotes the spread of AMR through horizontal gene transfer.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Abbreviations
- AMR:
-
Antimicrobial Resistance
- MAR:
-
Multiple Antibiotic Resistance Index
- NCBI:
-
National Center for Biotechnology Information
- BLAST:
-
Basic Local Alignment Search Tool
- RDP:
-
Ribosomal Database Project
- MIC:
-
Minimum Inhibitory Concentration
- AO:
-
Acridine Orange
- ARG:
-
Antibiotic Resistance Gene
- ATCC:
-
American Type Culture Collection
References
Alcaide E, Herraiz S, Esteve C (2006) Occurrence of Edwardsiella tarda in wild European eels Anguilla anguilla from Mediterranean Spain. Dis Aquat Org 73:77–81
Algammal AM, Mabrok M, Eat M, Alfifi KJ, Esawy AM, Elmasry N, El-Tarabili R (2021) Prevalence, antimicrobial resistance (AMR) pattern, virulence determinant and AMR genes of emerging multi-drug resistant Edwardsiella tarda in Nile tilapia and African catfish. Aquaculture 548:737643. https://doi.org/10.1016/j.aquaculture.2021.737643
Austin B, Austin DA (2016) Bacterial fish pathogens: disease of farmed and wild fish, 6th edn. Springer, Dordrecht
Barrow GI, Feltham RKA (2004) Cowan and Steel’s manual for the identification of medical bacteria, 3rd edn. Cambridge University Press, Cambridge
Chinabut S, Roberts RJ, Willoughby LG, Pearson MD (1995) Histopathology of snakehead, channa striatus (bloch), experimentally infected with the specific aphanomyces fungus associated with epizootic ulcerative syndrome (eus) at different temperatures. J Fish Dis 18:41–47. https://doi.org/10.1111/j.1365-2761.1995.tb01264.x
CLSI (2020) Methods for antimicrobial broth dilution and disk diffusion susceptibility testing of bacteria isolated from aquatic animals. 2nd ed. CLSI guideline VET03. Clinical and Laboratory Standards Institute, Wayne
Davies YM, de Oliveira MGX, Cunha MPV, Franco LS, Santos SLP et al (2018) Edwardsiella tarda outbreak affecting fishes and aquatic birds in Brazil. Vet Q 38 99–105. https://doi.org/10.1080/01652176.2018.1540070
Dharmaratnam A, Swaminathan T R, Kumar R, Basheer VS (2018) Aeromonas hydrophila associated with mass mortality of adult goldfish Carassius auratus in ornamental farms in India. Indian J Fish 65:116–126. https://doi.org/10.21077/ijf.2018.65.4.72719-14
Dubey S, Maiti B, Kim -H et al (2018) Genotypic and phenotypic characterization of Edwardsiella isolates from different fish species and geographical areas in Asia: Implications for vaccine development. J Fish Dis 42:1–16
Ewing WH, McWhorter AC, Escobar MR, Lubin AH (1965) Edwardsiella, a new genus of Enterobacteriaceae based on a new species, E. tarda. Int Bull Bacteriol Nomen Taxon 15:33–38. https://doi.org/10.1099/00207713-15-1-33
Hodgkinson JW, Ge J-Q, Grayfer L, Stafford J, Belosevic M (2012) Analysis of the immune response in infections of the goldfish (Carassius auratus L.) with Mycobacterium marinum. Dev Comp Immunol 38:456–465
Humphrey JD, Lancaster C, Gudkovs N, McDonald W (1986) Exotic bacterial pathogens Edwardsiella tarda and Edwardsiella ictaluri from imported ornamental fish Betta splendens and Puntius conchonius, respectively: isolation and quarantine significance. Aust Vet J 63:69–71
Jeremi S, Jaki-Dimi D, Veljovi L (2003) Citrobacter freundii as a cause of disease in fish. Acta Vet 53:399–410
Jin S, Fu S, Li R, Dang H, Gao D, Ye S, Jiang Z (2020) Identification and histopathological and pathogenicity analysis of Aeromonas salmonicida salmonicida from goldfish (Carassius auratus) in North China. Aquac Fish 5:36–41. https://doi.org/10.1016/j.aaf.2019.04.004
Kerie Y, Nuru A, Abayneh T (2019) Edwardsiella species infection in fish population and its status in Ethiopia. Fish Aqua J 10:266
Kumar R, Swaminathan TR, Arathi D, Basheer VS, Jena JK (2015) Mass mortality in ornamental fish, Cyprinus carpio koi caused by a bacterial pathogen, Proteus hauseri. Acta Trop 149:128–134. https://doi.org/10.1016/j.actatropica.2015.05.022
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA x: Molecular evolutionary Genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Krumperman PH (1983) Multiple antibiotic resistance indexing of Escherichia coli to identify high risk sources of fecal contamination of foods. Appl Environ Microbiol 46:165–170
Leung KY, Siame BA, Tenkink BJ, Noort RJ, Mok YK (2012) Edwardsiella tarda - virulence mechanisms of an emerging gastroenteritis pathogen. Microbes Infect 14:26–34
Mohanty BR, Sahoo PK (2007) Edwardsiellosis in fish: a brief review. J Biosci 32:1331–1344
Miller SA, Dyke DD, Polesk HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acid Res 16:12–15
Miyazaki T, Egusa S (1976) Histopathological studies of edwardsiellosis of the Japanese eel (Anguilla japonica), 1: suppurative interstitial nephritis form. Fish Pathol 11:33–43
Niu G, Wongsathein D, Boonyayatra S, Khattiya R (2019) Occurrence of multiple antibiotic resistance and genotypic characterization in Edwardsiella tarda isolated from cage-cultured hybrid red tilapia (Oreochromis sp.) in the Ping River, Northern Thailand. Aquacult Res 50:3643–3652
Park SB, Aoki T, Jung TS (2012) Pathogenesis of and strategies for preventing Edwardsiella tarda infection in fish. Vet Res 43:67. doi: https://doi.org/10.1186/1297-9716-43-67
Preena PG, Dharmarathnam A, Raj NS, Kumar TVA, Raja SA, Nair RR, Swaminathan TR (2019) Diversity of antimicrobial resistant pathogens from a freshwater ornamental fish farm. Lett Appl Microbiol 71:108–116
Preena PG, Swaminathan TR, Kumar VJR, Singh ISB (2020a) Antimicrobial resistance in aquaculture: a crisis for concern. Biologia 75:1497–1517
Preena PG, Dharmarathnam A, Raj NS, Raja SA, Nair RR, Swaminathan TR (2020b) Antibiotic resistant Enterobacteriaceae from diseased freshwater goldfish. Arch Microbiol 203:219–231
Raj NS, Swaminathan TR, Dharmaratnam DA, Raja SA, Ramraj D, Lal KK (2019) Aeromonas veronii caused bilateral exophthalmia and mass mortality in cultured Nile tilapia, Oreochromis niloticus (L.) in India. Aquaculture 512:734278
Reed MJ, Muench M (1938) A simple method for estimating 50% endpoints. Am J Trop Med Hyg 27:493–497. https://doi.org/10.1093/oxfordjournals.aje.a118408
Reichley SR, Ware C, Steadman J, Gaunt PS, García JC et al (2017) Comparative phenotypic and genotypic analysis of Edwardsiella isolates from different hosts and geographic origins, with emphasis on isolates formerly classified as E. tarda, and evaluation of diagnostic methods. J Clin Microbiol 55:3466–3491. https://doi.org/10.1128/jcm.00970-17
Sakai T, Kanai K, Osatomi K, Yoshikoshi K (2003) Identification of a 19.3-kDa protein in MRHA-positive Edwardsiella tarda: putative fimbrial major subunit. FEMS Microbiol Lett 226:127–133
Sakai T, Kanai K, Osatomi K, Yoshikoshi K (2004) Identification and characterization of fimbrial gene cluster of Edwardsiella tarda expressing mannose-resistant hemagglutination. Fish Pathol 39:87–93. https://doi.org/10.3147/jsfp.39.87
Sakai T, Lida T, Osatomi K, Kanai K (2007) Detection of type 1 fimbrial genes in fish pathogenic and nonpathogenic Edwardsiella tarda strains by PCR. Fish Pathol 42:115–117
Sakai T, Matsuyama T, Sano M, Iida T (2009) Identification of novel putative virulence factors, adhesin AIDA and type VI secretion system, in atypical strains of fish pathogenic Edwardsiella tarda by genomic subtractive hybridization. Microbiol Immunol 253:131–139. https://doi.org/10.1111/j.1348-0421.2009.00108.x
Shen Y-R, Chen J-D (2005) Expression of virulent genes of Edwardsiella tarda correlated with mortality of diseased-fish infection. J Fish Soc Taiwan 32:80–81
Shetty M, Maiti B, Venugopal M, Karunasagar I, Karunasagar I (2014) First isolation and characterization of Edwardsiella tarda from diseased striped catfish, Pangasianodon hypophthalmus (Sauvage). J Fish Dis 37:265–271. https://doi.org/10.1111/jfd.12039
Stock I, Wiedemann B (2001) Natural antibiotic susceptibilities of Edwardsiella tarda, E. ictaluri, and E. hoshinae. Antimicrob Agents Chemother 45:2245-2255. https://doi.org/10.1128/AAC.45.8.2245-2255.2001
Tan YP, Zheng J, Tung SL, Rosenshine I, Leung KY (2005) Role of type III secretion in Edwardsiella tarda virulence. Microbiology 151:2301–2313. https://doi.org/10.1099/mic.0.28005-0
Trabulsi L, Ewing W (1962) Sodium acetate medium for the differentiation of Shigella and Escherichia cultures. Public Health Lab 20:137–140
Verjan N, Hirono I, Aoki T (2005) Genetic loci of major antigenic protein genes of Edwardsiella tarda. Appl Environ Microbiol 71:5654–5658
Verma DK, Rathore G, Pradhan PK, Sood N, Punia P (2015) Isolation and characterization of Flavobacterium columnare from freshwater ornamental goldfish Carassius auratus. J Environ Biol 36:43–49
Walczak N, Puk K, Guz L (2017) Bacterial flora associated with diseased freshwater ornamental fish. J Vet Res 61:445–449
Wang X, Wang Q, Xiao J, Liu Q, Wu H, Zhang Y (2010a) Hemolysin EthA in Edwardsiella tarda is essential for fish invasion in vivo and in vitro and regulated by two-component system EsrA-EsrB and nucleoid protein HhaEt. Fish Shellfish Immunol 29:1082–1091
Wang B, Mo LZ, Xiao P, Li J, Zou YX, Hao B, Li GY (2010b) EseD, a putative T3SS translocon component of Edwardsiella tarda, contributes to virulence in fish and is a candidate for vaccine development. Mar Biotechnol 12:678–685
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal dna amplification for phylogenetic study. J Bacteriol 173:697–703
Xu T, Zhang X-H (2014) Edwardsiella tarda: an intriguing problem in aquaculture. Aquaculture 431:129–135. https://doi.org/10.1016/j.aquaculture.2013.12.001
Yu JE, Cho MY, Kim J-W, Kang HY (2012) Large antibiotic-resistance plasmid of Edwardsiella tarda contributes to virulence in fish. Microb Pathog 52:259–266
Acknowledgements
We kindly acknowledge Department of Veterinary Pathology, Veterinary College and Research Institute, Namakkal, Tamil Nadu for carrying out the histopathological studies. The first author thanks Department of Science and Technology (SERB) for providing National Postdoctoral Research fellowship (PDF/2017/000378) and Director, ICAR-National Bureau of Fish Genetic Resources, Lucknow for the support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest to declare.
Ethics approval
All the experimental challenge procedures in this study (Proposal number: NBFGR/IAEC/2019/0014) were evaluated and approved by Institute Animal ethics Committee (IAEC) of ICAR National Bureau of Fish Genetic Resources (NBFGR) (CPCESA Registration No: 909/GO/Re/S/05/CPCSEA dated 09.09.2005 and CPCSEA Ref file No. 25/111/2014-CPCESA dated 05th December 2018) vide approval Number G/IAEC/2019/1 dated 04th October 2019.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Preena, P.G., Dharmaratnam, A. & Swaminathan, T.R. A peek into mass mortality caused by antimicrobial resistant Edwardsiella tarda in goldfish, Carassius auratus in Kerala. Biologia 77, 1161–1171 (2022). https://doi.org/10.1007/s11756-022-01007-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11756-022-01007-9