Abstract
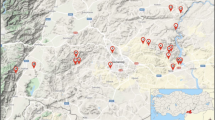
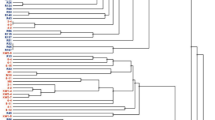
In this study, the diversity of root-nodulating bacteria associated with Pisum sativum L. cultivated in the Eastern and Western Black Sea regions (11 provinces) and three provinces in the Marmara region were investigated. rDNA-ITS RFLP analysis revealed nine groups within the 40 isolates collected in the study. Based on the phylogenetic analysis of the recA, atpD and glnII genes, isolates within the RFLP groups were assigned to the following Rhizobium species; Group-I: R. laguerreae; Groups -II and -IV: R. ruizarguesonis; Groups -III, -V, -VII, -VIII: genospecies-B (gsB) of the R. leguminosarum complex (Rlc); Group-VI: Rlc gsE; and Group-IX: Rlc gsA. From the nodC-RFLP analysis and phylogenetic analysis of the nodC gene, all rhizobial isolates obtained in this study were assigned to the symbiovar. viciae variety of the related Rhizobium species. In this study, Rlc gsB (n: 19) was the species that most commonly caused the nodulation of P. sativum in the northern Anatolia, followed by R. ruizarguesonis (n: 8). Rhizobium laguerreae (n: 5) was a widespread but not common species, and Rlc gsA (n: 3) and Rlc gsE (n: 5) had a distribution limited to the Eastern and Western Black Sea regions, respectively. Overall, results obtained from species diversity analysis, nodC phylogenetic analysis and nodX analysis suggest a common phylogeographic history for pea-nodulating rhizobia in northern Anatolia and western Europe. Separately, this study presents the first reports of R. laguerreae and R. ruizarguesonis from P. sativum root nodules in Turkey and the first report of R. ruizarguesonis outside of Europe.



Similar content being viewed by others
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author upon reasonable request.
Code availability
Not applicable.
Abbreviations
- AIC:
-
Akaike’s Information Criterion
- ANI:
-
Average Nucleotide Identities
- BIC:
-
Bayesian Information Criterion
- BLAST:
-
Basic Local Alignment Search Tool
- BNF:
-
Biological nitrogen fixation
- Gs:
-
Genospecies
- HUP+ :
-
Hydrogen Uptake Positive
- ML:
-
Maximum Likelihood
- MLSA:
-
Multilocus Sequence Analysis
- MP:
-
Maximum Parsimony
- NCBI:
-
National Center for Biotechnology Information
- NJ:
-
Neighbor-Joining
- pSym :
-
Symbiotic plasmid
- RFLP:
-
Restriction Fragments Length Polymorphi
- Rlc:
-
R. leguminosarum species Complex
- TBE:
-
Tris Borate EDTA
- TBR:
-
Tree Bisection and Re-connection
- YMA:
-
Yeast Extract Mannitol Agar
References
Anonymous TMO (2018) yılı bakliyat sektör raporu. Toprak mahsülleri ofisi genel müdürlüğü. Ankara p-22. https://doi.org/10.1007/s11756-021-00831-9
Basbuga S, Basbuga S, Yayla F, Mahmoud AM, Can C (2021) Diversity of rhizobial and non-rhizobial bacteria nodulating wild ancestors of grain legume crop plants. Int Microbiol. https://doi.org/10.1007/s10123-020-00158-6
Bastianelli D, Grosjean F, Peyronnet C, Duparque M, Regnier JM (1998) Feeding value of pea (Pisum sativum, L.) 1. Chemical composition of different categories of pea. Anim Sci 67:609–619. https://doi.org/10.1017/S1357729800033051
Bayraklı B, Dengiz O (2020) An evaluation of heavy metal pollution risk in tea cultivation soils of micro–catchments using various pollution indexes under humid environmental condition. Rend Lincei Sci Fis Nat 31:393–409. https://doi.org/10.1007/s12210-020-00901-1
Belhadi D, de Lajudie P, Ramdania N et al (2018) Vicia faba L. in the Bejaia region of Algeria is nodulated by Rhizobium leguminosarum sv. viciae, Rhizobium laguerreae and two new genospecies. Sys App Microbiol 41:122–130. https://doi.org/10.1016/j.syapm.2017.10.004
Brewin NJ, Beringer JE, Johnston AWB (1980) Plasmid-mediated trensfer of host-range specificity between two strains of Rhizobium leguminosarum. J Gen Microbiol 120:413–420. https://doi.org/10.1099/00221287-120-2-413
Cavassim MIA, Moeskjær S, Moslemi C, Fields B, Bachmann A, Vilhjalmsson BJ, Schierup MH, Young JPW, Andersen SU (2020) Symbiosis genes show a unique pattern of introgression and selection within a Rhizobium leguminosarum species complex. Microbial Genomics 6:1–15. https://doi.org/10.1099/mgen.0.000351
Coskun M, Cayir A, Coskun M, Kilic O (2011) Heavy metal deposition in moss samples from East and South Marmara Region, Turkey. Environ Monit Assess 174:219–227. https://doi.org/10.1007/s10661-010-1452-1
Cousin R (1997) Peas (Pisum sativum L.). Field Crop Res 53:111–130. https://doi.org/10.1016/S0378-4290(97)00026-9
Davis EO, Evans IJ, Johnston AWB (1988) Identification of nodX, a gene that allows the Rhizobium leguminosarum biovar viciae strain TOM to nodulate Afghanistan peas. Mol Gen Genet 212:531–535. https://doi.org/10.1007/BF00330860
Flores-Félix JD, Sánchez-Juanes F, García-Fraile P, Valverde A, Mateos PF, Gonzalez-Buitrago JM, Velazquez E, Rivas R (2019) Phaseolus vulgaris is nodulated by the symbiovar viciae of several genospecies of Rhizobium laguerreae complex in a Spanish region where Lens culinaris is the traditionally cultivated legume. Syst Appl Microbiol 42:240–247. https://doi.org/10.1016/j.syapm.2018.10.009
Flores-Félix JD, Carro L, Cerda-Castillo E, Squartini A, Rivas R, Velázquez E (2020) Analysis of the interaction between Pisum sativum L. and Rhizobium laguerreae strains nodulating this Legume in Northwest Spain. Plants 9:1755. https://doi.org/10.3390/plants9121755
Fred EB, Baldwin IL, McCoy E (1932) Root nodule bacteria and leguminous plants. University of Wisconsin Stud Sci 5, Madison
Gaunt MW, Turner SL, Rigottier-Gois L, Lloyd-Macgilp SA, Young JP (2001) Phylogenies of atpD and recA support the small subunit rRNA-based classification of rhizobia. Int J Syst Evol Microbiol 51:2037–2048. https://doi.org/10.1099/00207713-51-6-2037
Glaeser SP, Kampfer P (2015) Multilocus sequence analysis (MLSA) in prokaryotic taxonomy. Syst Appl Microbiol 38:237–245. https://doi.org/10.1016/j.syapm.2015.03.007
Graham PH, Sadowsky MJ, Kersters HH, Barnet YM, Bradley RS, Cooper JE, De Ley DJ, Jarvis BDW, Roslycky EB, Strijdom BW, Young JPW (1991) Proposed minimal standards for the description of new genera and species of root-and stem-nodulating bacteria. Int J Syst Bacteriol 41:582–587. https://doi.org/10.1099/00207713-41-4-582
Guindon S, Gascuel O (2003) A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704. https://doi.org/10.1080/10635150390235520
Gürkanli CT, Özkoç İ, Gündüz İ (2014) Genetic diversity of Vicia faba L. and Pisum sativum L. nodulating rhizobia in the central Black Sea region of Turkey. Ann Microbiol 64:99–112. https://doi.org/10.1007/s13213-013-0638-5
Gürkanlı CT, Özkoç İ, Gündüz İ (2013) Genetic diversity of rhizobia nodulating common bean (Phaseolus vulgaris L.) in the Central Black Sea region of Turkey. Ann Microbiol 63:971–987. https://doi.org/10.1007/s13213-012-0551-3
Gürkanlı CT, Özkoç İ, Kıvanç M (2018) First report of two new rhizobial species nodulating Phaseolus vulgaris L. in Turkey. BioDiCon 11:106–114
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acid S 41:95–98
Herridge DF, Peoples MB, Boddey RM (2008) Global inputs of biological nitrogen fixation in agricultural systems. Plant Soil 311:1–18. https://doi.org/10.1007/s11104-008-9668-3
Hirsch AM, Lum MR, Downie JA (2001) What makes the Rhizobia-Legume symbiosis so special? Plant Physiol 127:1484–1492. https://doi.org/10.1104/pp.010866
Holl FB (1975) Host plant control of the inheritance of dinitrogen fixation in the Pisum-Rhizobium symbiosis. Euphytica 24:767–770. https://doi.org/10.1007/BF00132916
Janzen J, Brester G, Smith V (2014) Dry peas: trends in production, trade, and price. Agricultural Marketing Policy Center, briefing N°57, 7p
Jorrin B, Palacios JM, Peix A, Imperial J (2020) Rhizobium ruizarguesonis sp. nov., isolated from nodules of Pisum sativum L. Syst Appl Microbiol 43:126090. https://doi.org/10.1016/j.syapm.2020.126090
Karayel R, Bozoğlu H (2008) Türkiye’nin farklı bölgelerinden toplanan yerel bezelye popülasyonunun bazı agronomik özellikleri. OMÜ Zir Fak Dergisi 23:32–38
Keskin Ş (2018) Melen nehri sedimentlerinin jeokimyasal özellikleri (Düzce ovası). ÖHÜ Müh Bilim Derg 7:1072–1077
Kozik A, Heidstra R, Horvath B et al (1995) Pea lines carrying sym1 or sym2 can be nodulated by Rhizobium strains containing nodX; sym1 and sym2 are allelic. Plant Sci 108:41–49. https://doi.org/10.1016/0168-9452(95)04123-C
Kumar N, Lad G, Giuntini E, Kaye ME, Udomwong P, Shamsani NJ, Young JPW, Bailly X (2015) Bacterial genospecies that are not ecologically coherent: population genomics of Rhizobium leguminosarum. Open Biol 5:140133. https://doi.org/10.1098/rsob.140133
Kumar N, Pandey S, Mishra S, Mishra DP, Pandey VP (2019) Studies genetic divergence for yield and its component traits in pea (Pisum sativum L.) in sodic condition. J Pharmacogn Phytochem 8:302–304
Kwon SW, Park JY, Kim JS, Kang JW, Cho YH, Lim CK, Parker MA, Lee GB (2005) Phylogenetic analysis of the genera Bradyrhizobium, Mesorhizobium, Rhizobium and Sinorhizobium on the basis of 16S rRNA gene and internally transcribed spacer region sequences. Int J Syst Evol Microbiol 55:263–270. https://doi.org/10.1099/ijs.0.63097-0
Laguerre G, Nour SM, Macheret V, Sanjuan J, Drouin P, Amarger N (2001) Classification of rhizobia based on nodC and nifH gene analysis reveals a close phylogenetic relationship among Phaseolus vulgaris symbionts. Microbiology + 147:981–993. https://doi.org/10.1099/00221287-147-4-981
Lie TA (1978) Symbiotic specialization in pea plants: the requirement of specific Rhizobium strains for peas from Afghanistan. Ann Appl Biol 88:462–465. https://doi.org/10.1111/j.1744-7348.1978.tb00743.x
Ma SW, Iyer VN (1990) New field isolates of Rhizobium leguminosarum bivar viciae that nodulate the primitive pea cultivar Afghanistan in addition to modern cultivars. Appl Environ Microb 56:2206–2212. https://doi.org/10.1128/AEM.56.7.2206-2212.1990
Martens M, Dawyndt P, Coopman R, Gillis M, De Vos P, Willems A (2008) Advantages of multilocus sequence analysis for taxonomic studies: a case studyusing 10 housekeeping genes in the genus Ensifer (including former Sinorhizobium). Int J Syst Evol Microbiol 58:200–214. https://doi.org/10.1099/ijs.0.65392-0
Martínez-Molina E, Sánchez Juanes F, Carro L, Flores-Felix JD, Martinez-Hidalgo P, Castillo EC, Buitrago JMG, Velazquez E (2016) Identification of rhizobial strains nodulating Pisum sativum in Northern Spain soils by MALDI-TOF MS (Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry) analysis. In: González-Andrés F, James E (eds) Biological Nitrogen Fixation and Beneficial Plant-Microbe Interaction. Springer, Cham, pp 37–44
Mousavi SA, Willems A, Nesme X, de Lajudie P, Lindström K (2015) Revised phylogeny of Rhizobiaceae: Proposal of the delineation of Pararhizobium gen. nov., and 13 new species combinations. Syst Appl Microbiol 38:84–90. https://doi.org/10.1016/j.syapm.2014.12.003
Ohyama T (2017) The Role of Legume-Rhizobium Symbiosis in Sustainable Agriculture. In: Sulieman S, Tran LS (eds) Legume Nitrogen Fixation in Soils with Low Phosphorus Availability. Springer, Cham. https://doi.org/10.1007/978-3-319-55729-8_1
Ovtsyna AO, Rademaker GJ, Esser e, Weinman J, Rolfe BG, Tikhonovich IA, Lugtenberg BJ, Thomas-Oates JE, Spaink HP (1999) Comparison of characteristics of the nodX genes from various Rhizobium leguminosarum strains. Mol Plant Microbe In 12:252–258. https://doi.org/10.1094/MPMI.1999.12.3.252
Pehlivan E, Aslantaş R (2020) Detection and evaluation of some heavy metals (Cr, Ni, Pb) in hazelnut orchards located between Trabzon and Giresun. J Fac Eng Archit Gazi Univ 35:431–442
Peix A, Ramírez-Bahena MH, Velázquez E, Bedmar EJ (2015) Bacterial associations with legumes. CRC Crit Rev Plant Sci 34:1-3 17. https://doi.org/10.1080/07352689.2014.897899
Posada D (2008) jModel test: phylogenetic model averaging. Mol Biol Evol 25:1253–1256. https://doi.org/10.1093/molbev/msn083
Prakash RK, Schilperoot RA, Nuti MP (1981) Large plasmids of fast-growing rhizobia: homology studies and location of structural nitrogen fixation (nif) genes. J Bacteriol 145:1129–1136. https://doi.org/10.1128/JB.145.3.1129-1136.1981
Rahi P, Kapoor R, Young JPW, Gulati A (2012) A genetic discontinutity in root-nodulating bacteria of cultivated pea in the Indian trans-Himalayas. Mol Ecol 21:145–159. https://doi.org/10.1111/j.1365-294X.2011.05368.x
Rahi P, Giram P, Chaudhari D et al (2020) Rhizobium indicum sp. nov., isolated from root nodules of pea (Pisum sativum) cultivated in the Indian trans-Himalayas. Syst Appl Microbiol 43:126127. https://doi.org/10.1016/j.syapm.2020.126127
Ramirez-Bahena MH, Garcia-Fraile P, Peix A, Valverde A, Rivas R, Igual JM, Mateos PF, Martinez-Molina E, Velazquez E (2008) Revision of the taxonomic status of the species Rhizobium leguminosarum (Frank 1879) Frank 1889AL, Rhizobium phaseoli Dangeard 1926AL and Rhizobium trifolii Dangeard 1926AL. R. trifolii is a later synonym of R. leguminosarum. Reclassification of the strain R. leguminosarum DSM 30132 (= NCIMB 11478) as Rhizobium pisi sp. nov. Int J Syst Evol Microbiol 58:2484–2490. https://doi.org/10.1099/ijs.0.65621-0
Rashid HM, Gonzalez J, Young JPW, Wink M (2014) Rhizobium leguminosarum is the symbiont of lentils in the Middle East and Europe but not in Bangladesh. FEMS Microbiol Ecol 87:64–77. https://doi.org/10.1111/1574-6941.12190
Rogel MA, Ormeño-Orrillo E, Martinez Romero E (2011) Symbiovars in rhizobia reflect bacterial adaptation to legumes. Syst Appl Microbiol 34:96–104. https://doi.org/10.1016/j.syapm.2010.11.015
Saidi S, Ramirez-Bahena MH, Santillana N, Zuniga D, Alvarez-Martinez E, Peix A, Mhamdi R, Velazquez E (2014) Rhizobium laguerreae sp. nov. nodulates Vicia faba on several continents. Int J Syst Evol Microbiol 64:242–247. https://doi.org/10.1099/ijs.0.052191-0
Santillana N, Ramirez-Bahena MH, Garcia-Fraile P, Velazquez E, Zuniga D (2008) Phylogenetic diversity based on rrs, atpD, recA genes and 16S-23S intergenic sequence analyses of rhizobial strains isolated from Vicia faba and Pisum sativum in Peru. Arch Microbiol 189:239–247. https://doi.org/10.1007/s00203-007-0313-y
Shamseldin A, Abdelkhalek A, Sadowsky MJ (2017) Recent changes to the classification of symbiotic, nitrogen-fixing, legume-associating bacteria: a review. Symbiosis 71:91–109. https://doi.org/10.1007/s13199-016-0462-3
Singh Z, Singh G (2018) Role of Rhizobium in chickpea (Cicer arietinum) production - A review. Agric Rev 39:31–39. https://doi.org/10.18805/ag.R-1699
Somasegaran P, Hoben HJ (1985) Methods in legume-rhizobium Technology. USAID, Washington, DC
Swofford DL (1998) PAUP* Phylogenetic analysis using parsimony (*and Other Methods). Version 4 beta 10. Sinauer Associates, Sunderland
Taha K, Berraho EB, El Attar I, Dekkiche S, Aurag J, Bena G (2018) Rhizobium laguerreae is the main nitrogen-fixing symbiont of cultivated lentil (Lens culinaris) in Morocco. Syst Appl Microbiol 41:113–121. https://doi.org/10.1016/j.syapm.2017.09.008
Tan Z, Hurek T, Vinuesa P, Müller P, Ladha JK, Reinhold-Hurek B (2001) Specific detection of Bradyrhizobium and Rhizobium Strains colonizing Rice (Oryza sativa) roots by 16S-23S ribosomal DNA intergenic spacer-targeted PCR. App Environ Microbial 67:3655–3664. https://doi.org/10.1128/AEM.67.8.3655-3664.2001
Temizkan G, Arda N (2004) Moleküler biyolojide kullanılan yöntemler. Nobel Tıp Kitabevleri, İstanbul
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX-Windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882. https://doi.org/10.1093/nar/25.24.4876
Van Berkum P, Beyene D, Vera FT, Keyser HH (1995) Variability among Rhizobium strains originating from nodules of Vicia faba. Appl Environ Microbiol 61:2649–2653. https://doi.org/10.1128/AEM.61.7.2649-2653.1995
Verma A, Rawat AK, More N (2014) Extent of nitrate and nitrite pollution in ground water of rural areas of Lucknow. Curr World Environ 9:114–122
Vincent JM (1970) A manuel for the practical study of the root-nodule bacteria. Blackwell Scientific Publications, Oxford
Vinuesa P, Silva C, Werner D, Martinez-Romero E (2005) Population genetics and phylogenetic ınference in bacterial molecular systematics: the roles of migration and recombination in Bradyrhizobium species cohesion and delineation. Mol Phylogenet Evol 34:29–54. https://doi.org/10.1016/j.ympev.2004.08.020
Wang ET, Chen WF, Tian CF, Young JPW, Chen WX (2019) Ecology and evolution of rhizobia principles and applications. Springer Nature Singapore Pte Ltd. https://doi.org/10.1007/978-981-32-9555-1
Young JPW, Moeskjær S, Afonin A et al (2020) Defining the Rhizobium leguminosarum species complex. Preprints 2020120297. https://doi.org/10.20944/preprints202012.0297.v1
Zahran HH, Chahboune R, Moreno S et al (2013) Identification of rhizobial strains nodulating Egyptian grain legumes. Int Microbiol 16:157–163. https://doi.org/10.2436/20.1501.01.190
Zhang YJ, Zheng WT, Everall I, Young JPW, Zhang XX, Tian CF, Sui XH, Wang ET, Chen WX (2015) Rhizobium anhuiense sp. nov., isolated from effective nodules of Vicia faba and Pisum sativum. Int J Syst Evol Microbiol 65:2960–2967. https://doi.org/10.1099/ijs.0.000365
Zohary D, Hopf M, Weiss E (2012) Domestication of plants in the old World. Oxford Univ Press Inc, New York
Acknowledgements
The author thanks Gregory T. Sullivan for editing an earlier version of this manuscript.
Funding
The author did not receive support from any organization for the submitted work.
Author information
Authors and Affiliations
Contributions
All aspects of this work were completed by the author.
Corresponding author
Ethics declarations
Consent to participate
This study was completed by one author and thus consent to participate is not applicable.
Consent to publication
This study was completed by one author and thus consent to publication is not applicable.
Conflicts of interest/Competing interests
The author has no conflicts of interest to declare that are relevant to the content of this article.
Ethics approval
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX .53 MB)
Rights and permissions
About this article
Cite this article
Gürkanlı, C.T. Genetic diversity of rhizobia associated with Pisum sativum L. in the Northern part of Turkey. Biologia 76, 3149–3162 (2021). https://doi.org/10.1007/s11756-021-00831-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11756-021-00831-9