Abstract
Delta (Δ) 5 desaturase is a key enzyme for the biosynthesis of health-beneficial long chain polyunsaturated fatty acids such as arachidonic acid (ARA, C20:4n-6), eicosapentaenoic acid (C20:5n-3) and docosahexaenoic acid (C22:6n-3) via the “desaturation and elongation” pathways. A full length Δ5 desaturase gene from Euglena gracilis (EgΔ5D) was isolated by cloning the products of polymerase chain reaction with degenerate oligonucleotides as primers, followed by 5′ and 3′ rapid amplification of cDNA ends. The whole coding region of EgΔ5D was 1,350 nucleotides in length and encoded a polypeptide of 449 amino acids. BlastP search showed that EgΔ5D has about 39 % identity with a Δ5 desaturase of Phaeodactylum tricornutum. In a genetically modified dihomo-gamma-linoleic acid (DGLA, C20:3n-6) producing Yarrowia lipolytica strain, EgΔ5D had strong Δ5 desaturase activity with DGLA to ARA conversion of more than 24 %. Functional dissection of its HPGG and HDASH motifs demonstrated that both motifs were important, but not necessary in the exact form as encoded for the enzyme activity of EgΔ5D. A double mutant EgΔ5D-34G158G with altered sequences within both HPGG and HDASH motifs was generated and exhibited Δ5 desaturase activity similar to the wild type EgΔ5D. Codon optimization of the N-terminal region of EgΔ5D-34G158G and substitution of the arginine with serine at residue 347 improved substrate conversion to 27.6 %.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
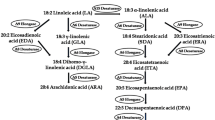
There is increasing interest in the recognized health benefits of long chain polyunsaturated fatty acids (LC-PUFA) such as arachidonic acid (ARA, C20:4n-6), eicosapentaenoic acid (EPA, C20:5n-3) and docosahexaenoic acid (DHA, C22:6n-3) for both humans and animals [1–3]. Since mammals lack delta (Δ) 12- and Δ15-desaturases, ARA, EPA and DHA cannot be synthesized de novo and must be obtained either in the diet or synthesized through “desaturation and elongation” pathways (Fig. 1) from essential fatty acids linoleic acid (LNA, 18:2n-6) and/or alpha (α)-linolenic acid (ALA 18:3n-3). These LC-PUFA are important fatty acids for human growth and development. For example, ARA, a precursor of EPA, is abundant in the brain and muscles. As a lipid second messenger ARA is involved in cellular signaling and is a key inflammatory intermediate [3]. EPA is a precursor of DHA, and induces a broad anti-inflammatory response [1–3]. DHA is a major ω-3 fatty acid in the mammalian central nervous system and enhances synaptic activities in neuronal cells [1–4]. EPA and DHA are the precursors of E- and D-series resolvins, respectively. These two classes of resolvins have distinct structural, biochemical and pharmacological properties [5, 6]. ARA and DHA together play critical roles for neurological development and health [4, 7]. Dietary EPA and DHA can effectively reduce the level of blood triglycerides in human [8]. Increased intake of EPA-rich supplement has beneficial effects on coronary heart disease, high blood pressure, inflammatory disorders and mental illness [9, 10].
Currently, the primary source of EPA and DHA is marine fish oil. Most of the EPA and DHA in fish oil are from their cold-water oceanic microalgae food sources. More than 85 % of isolated fish oil is used for aquaculture. In the case of salmon-farming, fish oils from approximately 4 pounds of fish are needed to raise one pound of salmon filet, the fish-in and fish-out ratio is about 4:1. In today’s environment, wild-caught fish often contain contaminants such as methylmercury, polychlorinated biphenyls, dioxins and several other halogenated persistent organic pollutants [11]. With ever-growing human populations, and limited sources of ocean fish, there is growing concern about the quality, quantity and sustainability of fish oil.
In the last two decades, great efforts have been focused on developing different hosts for production of LC-PUFA. Wild type Mortierella alpina has been developed for commercial production of ARA [12], while Crypthecodinium cohnii and Schizochytrium have been developed for commercial production of DHA [13]. The ARA and DHA oils produced from these organisms have been largely used in infant formulas. Yarrowia lipolytica has been genetically engineered to contain an EPA biosynthesis pathway [14] allowing for the commercial production of EPA oil, NewHarvest™ (http://www.newharvest.com). The EPA oil has been used as a human nutritional supplement. Additionally, EPA-rich Yarrowia biomass has been used to feed a brand of farmed salmon, Verlasso™ (http://www.verlasso.com), with a fish-in and fish-out ratio of about 1:1. However, the current production scale and cost of ARA, EPA and DHA cannot meet the market demand.
Various organisms use different pathways to synthesize ARA, EPA and DHA. Crypthecodinium and Schizochytrium synthesize EPA and DHA through a polyketide-based pathway [15], while some species of algae, fungi, and protists synthesize ARA, EPA and DHA through fatty acid “desaturation and elongation” pathways [16, 17]. All “desaturation and elongation” pathways (Fig. 1) require LNA and/or ALA as a substrate, followed by the “Δ9 elongase and Δ8 desaturase” pathway or the “Δ6 desaturase and C18/20 elongase” pathway to synthesize ARA, EPA and DHA by orchestrated elongation and desaturation reactions. So far, genetic engineering has mainly employed these “desaturation and elongation” pathways to modify hosts such as plants and yeast to produce ARA, EPA and DHA [14, 18–23]. Production of ARA, EPA and DHA through “desaturation and elongation” pathways requires gene expression of Δ5 desaturase to catalyze the conversion of di-homo-γ-linolenic acid (DGLA, C20:3n-6) to ARA, with a similar activity of converting eicosatetraenoic acid (ETA, C20:4n-3) to EPA. Since the isolation of Δ5 desaturase gene from Mortierella alpina (MaΔ5D, 24, 25), several Δ5 desaturase genes have been isolated [26], however, there is little research about their structure/function relationship. Additionally, more efficient Δ5 desaturase genes are needed to ensure engineered organisms may produce high levels of ARA, EPA and DHA.
Δ5 desaturases are known as “front-end” desaturases, wherein desaturation occurs between a pre-existing double bond and the carboxyl terminus of the fatty acid [26–29]. Like other desaturases, Δ5 desaturase is an iron-containing and membrane-bound enzyme that requires both molecular oxygen and an electron transfer to introduce double bonds into an existing acyl chain. Microsomal cytochrome b 5 serves as electron donor to desaturase enzymes [26, 29, 30]. Fatty acid desaturation can be carried out via concerted action of multiple enzymes including NADH reductase, the desaturase enzyme, and cytochrome b 5 reductase. Alternatively, many desaturases contain both a cytochrome b 5 domain and a desaturase domain. For example, Δ4, Δ5, Δ6 and Δ8 desaturases have a cytochrome b 5 domain at their N-terminus [26]; Δ9 desaturase has a cytochrome b 5 domain at its C-terminus [30]. The cytochrome b 5 domain of all desaturases has a heme-binding “HPGG” motif. Previous studies using molecular dynamics simulations suggest the “HPGG” motif is important for heme group assembly and desaturase function [31, 32].
The active site of desaturases has been characterized as a diiron cluster that is bound to the enzyme by three regions of highly conserved histidine-rich (His-rich) motifs [27, 30]. These three His-rich motifs H(X)3–4H, H(X)2–3HH, and H/Q(X)2–3HH are conserved among all front-end desaturases, and the eight histidine residues of these three motifs are essential for catalytic activity [33]. In the case of Δ5 desaturase from MaΔ5D [24, 25], the exact amino acid sequence of the first His-rich motif (H(X)3–4H) is HDASH, which has been suggested as one of the characteristics of Δ5 desaturases and necessary for its function to convert DGLA to ARA [34]. Recent studies find that several Δ5 desaturases (GenBank accession #s: AAL82631, AAL13311, AAL92562, AAM09687, CAJ07076) do not contain the exact HDASH sequence. Due to the important role of Δ5 desaturases in LC-PUFA biosynthesis, a detailed understanding of the functional significance of the conserved HPGG and HDASH motifs may contribute to improvements in hosts biologically engineered to produce commercially valuable LC-PUFA.
We report the isolation of a Δ5 desaturase gene from Euglena gracilis (EgΔ5D). Expression of EgΔ5D in a genetically modified DGLA producing Y. lipolytica strain revealed that EgΔ5D had strong Δ5 desaturase activity. Functional dissection of HPGG and HDASH motifs demonstrated that neither the HPGG nor the HDASH motif is necessary in the exact form as encoded for enzyme activity of EgΔ5D. Various mutants, within HPGG or HDASH motif alone, or within both HPGG and HDASH motifs, are functionally equivalent or have higher Δ5 desaturase activity than the wild type EgΔ5D. Codon optimization of the N-terminal region of a double mutant EgΔ5D-34G158G and substitution of the arginine with serine at residue 347 effectively improved the enzyme’s substrate conversion.
Materials and Methods
Strains, Media and Growth Conditions
E. gracilis was kindly provided by Dr. Richard Triemer of Michigan State University (East Lansing, MI). Euglena growth (EG) media (per liter): 1 g sodium acetate, 1 g of beef extract, 2 g of Bacto® tryptone and 2 g of Bacto® yeast extract in 970 mL of water. After filter sterilizing, 30 mL of soil–water supernatant was aseptically added. A 1-mL aliquot of E. gracilis culture was transferred into 250 ml of EG Medium in a 500-mL glass bottle. The cultures were grown at 23 °C with a 16 h light, 8 h dark cycle for 2 weeks with no agitation.
Y. lipolytica strain Y2224 is a 5-fluoroorotic acid (FOA) resistant mutant of wild type strain American Type Culture Collection (ATCC, Rockville, MD) #20362 (Fig. 2a) with a mutation in the URA3 gene (Genbank accession#: No. AJ306421). Y. lipolytica strain Y4036U (Zhu et al., unpublished data) is a genetically modified strain with a Leu − and Ura − phenotype, also originated from the wild type strain ATCC #20362. Strain Y4036U produced approximately 18 % DGLA of total fatty acids (Fig. 2b) and is composed of heterologous genes encoding Δ12 desaturase of Fusarium moniliforme [35]; C16/18 elongase of M. alpina [36]; Δ9 elongase of E. gracilis [37] and synthetic mutant of Δ8 desaturase [38] derived from E. gracilis. Minimal Media + Leucine (MMLeu), High Glucose Media (HGM) and YPD medium were used as required for Y. lipolytica strains and cultured at 30 °C. MMLeu (per liter): 20 g of glucose; 1.7 g yeast nitrogen base without amino acids or ammonium sulfate; 0.1 g proline; 0.1 g leucine; pH 6.1. HGM (per liter): 80 g glucose, 2.58 g KH2PO4, 5.36 g K2HPO4, pH 7.5. YPD medium (per liter): 10 g of yeast extract, 20 g of Bacto peptone, and 20 g of glucose. Agar plates were prepared by addition of 20 g/l agar to liquid media.
General Techniques for Molecular Biology
Recombinant DNA techniques were used according to standard methods [39, 40]. Site-directed mutagenesis was performed according to the manufacturer’s protocol (QuikChange™, Stratagene; San Diego, CA). When PCR or site-directed mutagenesis was involved in the generation of mutants and/or cloning, DNA was sequenced to verify that no additional mutations were introduced.
Total RNA was extracted from the E. gracilis cells using the RNA STAT-60™ reagent (Amsbio LLC., Lake forest, CA). 85 µg of mRNA was purified from 1 mg of total RNA using the mRNA Purification Kit (Amersham Biosciences, Piscataway, NJ). Synthesis of cDNA from the E. gracilis mRNA was carried out using the adapter primer AP of 3′-RACE kit from Invitrogen (Carlsbad, CA) and the Smart IV oligonucleotide of BD-Clontech Creator™ Smart™ cDNA library kit (Mississauga, ON, Canada) as primers. The reverse transcription was performed with Superscript II reverse transcriptase of Invitrogen.
PCR reactions were carried out in a 50 µl total volume comprising: PCR buffer (containing 10 mM KCl, 10 mM (NH4)2SO4, 20 mM Tris–HCl (pH 8.75), 2 mM MgSO4, 0.1 % Triton X-100), 100 µg/mL BSA, 200 µM each deoxyribonucleotide triphosphate, 10 pmol of each primer, 10 ng cDNA of E. gracilis and 1 µl of Taq DNA polymerase (Epicentre Technologies, Madison, WI). The thermocycler conditions were set for 35 cycles at 95 °C for 1 min, 56 °C for 30 s and 72 °C for 1 min, followed by a final extension at 72 °C for 10 min. The DNA band with expected size was isolated from a 1 % agarose gel and cloned into pGEM-T easy vector (Promega, Madison, WI.).
Modified 5′ and 3′ RACE techniques were used to obtain the full length EgΔ5D. The cDNA product from E. gracilis mRNA was used as template, and all the primers used in the 5′ and 3′ RACE are listed in Supplemental Table S1. Specifically, a gene specific primer ODMWP480 and a generic primer CDSIII 5′ were used in the first round of 5′ RACE. The PCR amplifications were carried out in a 50 µl total volume, comprising: 25 µl of LA Taq™ pre-mix (TaKaRa Bio Inc., Otsu, Shiga, 520-2193, Japan), 10 pmol of each primer and 1 µl of Taq DNA polymerase (Epicentre Technologies, Madison, WI). The thermocycler conditions were the same as described above. One micro liter of this product was directly used in a second amplification, which differed from the first only in that the primers used, ODMWP479 and the generic primer DNR CDS 5′ were internal to the first set of primers. As no translation initiation codon was found in the product of the first round of 5′ RACE, the entire modified 5′ RACE protocol was repeated using gene specific primers YL791 and YL792 as primers instead of the primers used in the first round.
A variation of a 3′ RACE technique was used to isolate the C-terminal fragment of EgΔ5D. The combinations of primers ODMW469 and AUAP, and then YL470 and AUAP were used in the initial amplification and second round reaction, respectively. The PCR reactions were the same as those described for the 5′ RACE.
Yarrowia Expression Vector and Transformation
Yarrowia expression vector pDMW367 contained autonomous replication sequence 18 [41] and a URA3 gene (Genbank accession#: No. AJ306421) of Y. lipolytica. It also contains a FBAIN::EgΔ5D:Pex20 chimeric gene. The FBAIN is a promoter derived from the fructose-bisphosphate aldolase gene (FBA1) of Y. lipolytica [42]. The EgΔ5D is the coding region of a wild type Δ5 desaturase of E. gracilis, in which the amino acid at position 347 is arginine. The Pex20 was a terminator sequence of PEX20 gene (Genbank accession#: AF054613) of Y. lipolytica. Transformation of Y. lipolytica strain Y4036U was carried out as described by Chen et al. [43].
Cultivation of Y. lipolytica Transformants and Fatty Acid Analysis by Gas Chromatography
The Yarrowia expression plasmid and its derivatives were used to transform strain Y4036U individually. Transformants from each transformation were streaked onto new MMLeu plates and kept in a 30 °C incubator for 2 days. Cells from streaked plates were cultivated in 24 well blocks with 3 mL MMLeu, and incubated for 2 days at 30 °C with shaking at 200 rpm. The cells were then collected by centrifugation and resuspended in 3 mL HGM. The cells were incubated another 5 days at 30 °C with shaking at 200 rpm.
Fatty acid methyl esters from 1 ml cell culture of Y. lipolytica or E. gracilis were prepared as described [44], except that the fatty acid methyl esters were extracted with 0.5 ml of heptane and separated by Agilent 7890A GC using hydrogen as carrier gas supplied by a hydrogen generator (Parker Hannifin, Cleveland, OH). The oven temperature was programmed from 200 to 240 °C at a rate of 25 °C/min. The proportion of each fatty acid was based on the integrated peak area of the corresponding fatty acid methyl ester as a percent relative to the sum of all integrated peaks as calculated by Agilent ChemStation Software.
Results
Isolation of Δ5 Desaturase Gene from E. gracilis
Fatty acid profile analyses showed that there were moderate amounts of EDA, DGLA, ARA, EPA, docosapentaenoic acid (DPA 22:5n-3) and DHA produced in E. gracilis (data not shown), confirming that there was a “Δ9 elongase/Δ8 desaturase” pathway in E. gracilis [16, 38, 45]. The genes encoding the Δ9 elongase, Δ8 desaturase [16, 38], Δ5 desaturase, Δ17 desaturase, C20/22 elongase and Δ4 desaturase [45] were responsible for the production of EDA, DGLA, ARA, EPA, DPA and DHA, respectively (Fig. 1).
After comparison of four Δ5 desaturase genes, PiΔ5D from Pythium irregulare [46], PmΔ5D from Phytophthora megasperma (Genbank accession #: CAD53323), PtΔ5D from Phaeodactylum tricornutum [47], and DdΔ5D from Dictyostelium discoideum (Genbank accession #: XP_640331) as well as two Δ8 desaturase genes, EgΔ8D from E. gracilis [16, 38] and PlΔ8D from Pavlova lutheri [48], two conserved regions, GHH(I/V)YTN and N(Y/F)Q(V/I)EHH (Fig. 3) were selected to design primers to amplify a portion of EgΔ5D. To reduce the degeneracy of the primers, four primers (Supplemental Table S1: 5-1A to 1D) were generated for conserved region 1 and four primers (Supplemental Table S1: 5-5AR to 5-5DR) for the anti-sense strand of conserved region 2. One DNA fragment amplified with primers 5-1B and 5-5DR was cloned into pGEM-T Easy vector to generate pT-F10-1. DNA sequence showed that a 590 bp insert of pT-F10-1 encoded an amino acid sequence with 38 % identity and 53 % similarity to the amino acid sequence of the Δ8-sphingolipid desaturase of Thalassiosira pseudonana (TsΔ8D, Genbank accession #: AAX14502), and 37 % identity and 52 % similarity with PtΔ5D [47]. These data suggested that the 590 bp DNA fragment might be a part of a desaturase gene of E. gracilis. This gene was designated as putative EgΔ5D.
Alignment of the conserved regions among some Δ5 and Δ8 desaturases. The amino acid sequence alignment was performed with Clustal W analysis (MegAlign™ program of DNASTAR software). Identical residues are shaded in black. PtΔ5D, Δ5 desaturase from P. tricornutum (29, GenBank accession #: AAL92562); PmΔ5D, Δ5 desaturase from P. megasperma (GenBank accession #: CAD53323); PiΔ5D, Δ5 desaturase from P. irregulare (30, GenBack accession #: AAL13311); DdΔ5D, Δ5 desaturase from D. discoideum (GenBack accession #: XP_640311); EgΔ8D, Δ8 desaturase from E. gracilis (24, GenBack accession #: AAD45877) and PvΔ8D, Δ8 desaturase from P. lutheri [48]
5′ and 3′ RACE techniques were used to extend the 590 bp region of the putative EgΔ5D (Fig. 4). A 797 bp DNA fragment with no translation initiation codon was isolated by the first round of 5′ RACE. This 797 bp DNA fragment had a 238 bp overlap with the 5′ end of the 590 bp fragment of pT-F10-1 and provided 559 bp of 5′ upstream sequence. A 273 bp DNA fragment was generated by the second round 5′ RACE experiment. This 273 bp DNA fragment had 253 bp overlap with the 5′ part of the 797 bp DNA fragment and provided 20 bp of 5′ upstream sequence. Seventeen [17] bp of the 20 bp encoded the N-terminal portion of the putative EgΔ5D, including the translation initiation codon. A 464 bp DNA sequence was identified by one round of 3′ RACE. The first 184 bp of the 464 bp fragment encoded the C-terminal coding region, including the translation stop codon, of the putative EgΔ5D.
Assembly of the 5′ region, the original 590 bp fragment and the 3′ region resulted in a 1,633 bp contig, comprising the complete coding region with additional untranslated 5′ and 3′ ends (Fig. 4). The coding region of the putative EgΔ5D is 1,350 bp in length and encodes a peptide of 449 amino acids. BlastP searches using the full length putative EgΔ5D as the query sequence showed that it shares 39 % identity and 56 % similarity with PtΔ5D [47]; 37 % identity and 55 % similarity with TsΔ8D (Genbank accession #: AAX14502). Amino acid sequence alignment performed with the Clustal W analysis (MegAlign™ program of DNASTAR software) showed that EgΔ5D has <30 % identity with some represent Δ5 desaturases such as IgΔ5D from I. galbana (AEA72469); MaΔ5D from M. alpina [24, 25, 34]; PiΔ5D [46]; OtΔ5D from O. tauri (Genbank accession #: XP_003082424) and TaΔ5D from T. aureum [49]. Further analyses showed that EgΔ5D has only 20 % and 25.5 % identity with EgΔ8D [16] and EgΔ4D [45] desaturases of E. gracilis, respectively. It was found that the PCR products for the full length coding region of putative EgΔ5D had two versions, both having identical nucleotide sequence except at base pair positions 1,039 and 1,041. This disagreement resulted in a codon change from CGA to AGC. As such, one PCR product indicated arginine at position 347, whereas the second indicated serine. It was hypothesized that this discrepancy was raised at the stage of PCR amplification or during cDNA generation.
Determination of Δ5 Desaturase Activity and Topology Model of EgΔ5D
To study the function of the putative EgΔ5D, plasmid pDMW367 was generated to express the EgΔ5D coding region under the control of the strong constitutive FBAIN promoter [42] from Y. lipolytica. The four restriction sites (i.e., BglII, EcoRI, HindIII and NcoI) inside the coding region of EgΔ5D in pDMW367 were removed by site-directed mutagenesis to generate pDMW367-M4 (Supplemental Fig. S1). The amino acid sequence of EgΔ5D is identical in pDMW367 and pDMW367-M4 constructs, and the amino acid at position 347 is an arginine.
The pDMW367 and pDMW367-M4 constructs were used to transform DGLA producing strain Y4036U. There was no ARA produced in the parent strain Y4036U (Fig. 2b), while there was about 3.7 % ARA and 11.5 % DGLA produced in transformants of Y4036U with either the pDMW367 or pDMW367-M4 construct (Fig. 2c). These results demonstrated that EgΔ5D indeed encodes a Δ5 desaturase. EgΔ5D could convert DGLA to ARA with a conversion of about 24.3 %. The conversion of DGLA to ARA was calculated according to the formula: ([ARA product]/[DGLA substrate + ARA product]). The pDMW367 and pDMW367-M4 constructs were also used to transform strain Y2224, a FOA resistant mutant of wild type strain ATCC#20362. There was no GLA produced in the transformants with either the pDMW367 or pDMW367-M4 construct (data not shown). These results demonstrated that EgΔ5D does not have Δ6 desaturase activity; it is not a bi-functional enzyme.
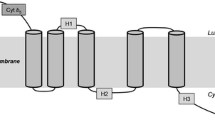
Like other fatty acid desaturases, EgΔ5D is also a membrane-bound enzyme and belongs to a super-family of membrane di-iron proteins with three His-rich motifs: HX(3, 4)H, HX(2, 3)HH and (H/Q)X(2, 3)HH. These His residues have been predicted to be located in the cytoplasmic face of the membrane and have been shown to be very important for enzyme activity [33]. Within EgΔ5D, these 3 His-rich motifs are the HDASH motif located from residue 155 to 159, the HIMRHH motif located from residue 190 to 195, and the QIEHH motif located from residue 385 to 389. The third His-rich motif contains a glutamine substitution that is common to other front-end desaturases. Based on transmembrane domain analysis (TMHMM Server v. 2.0, Center for Biological Sequence Analysis, BioCentrum-DTU, Technical University of Denmark, DK-2800 Lyngby, Denmark) and the location of the His-rich motifs, a topology model of EgD5D was developed (Fig. 5). The model shows that the N-terminal cytochrome b 5 domain is located in the cytosol. The topology model also predicts that EgΔ5D has a total of four transmembrane regions (amino acid residues 103–125, 130–152, 280–302 and 306–328) and two hydrophobic regions (amino acid residues 165–187, and 234–256). These hydrophobic segments are not membrane-spanning, and may represent hydrophobic patches located closed to the di-iron active site. Because the substrates for the desaturase is highly hydrophobic, they will likely partition into the lipid bilayer. Therefore we purport that the di-iron active site assembled from these His-clusters may occur at or very near the membrane surface.
Topology model of EgΔ5D. EgΔ5D is a membrane diiron protein with three His-rich motifs. The HX(3, 4)H motif, HDASH, is located from residues 155 to 159; the HX(2, 3)HH motif, HIMRHH, located from residues 190 to 195, and the (H/Q)X(2, 3)HH motif, QIEHH, located from residues 385 to 389. EgΔ5D has four trans-membrane domains (I–IV) and two hydrophobic stretches (residues from 165 to 187, and 234 to 256). Its N-terminal cytochrome b 5 domain and C-terminal region are located in the cytoplasm
The HPGG Motif is Important, but not Necessary for Δ5 Desaturase Activity of EgΔ5D
It has been suggested that the highly conserved HPGG motif plays a crucial role in heme group assembly, protein folding and stabilization in cytochrome b 5 proteins, with the histidine residue functioning as an axial heme ligand where a peptide chain reversal occurs [50]. Previous studies have demonstrated that the heme-binding HPGG motif, and in particular, the histidine residue, is essential for enzyme activity of desaturases with cytochrome b 5 domain [51–53]. Although sequence divergence in the vicinity of the HPGG motif is normal, the HPGG motif itself has been conserved throughout the evolution of all the Δ5 desaturase genes [54]. Thus it was claimed to be a characteristic of Δ5 desaturases and necessary for its function to convert DGLA to ARA [34].
To assess the functional significance of the HPGG motif (position 33–36) of EgΔ5D, we first elected the proline residue at position 34 (P34) as a target for amino acid substitution. Single amino acid mutations were carried out to generate all 19 amino acid substitution mutants (HxGG). Table 1 shows that the P34 residue could be substituted with several different amino acids without significantly impacting the Δ5 desaturase activity of EgΔ5D. The EgΔ5D-34A, EgΔ5D-34C, EgΔ5D-34K or EgΔ5D-34W mutants exhibited >90 % of the wild type EgΔ5D activity. The EgΔ5D-34G mutant was functionally equivalent to the wild type EgΔ5D.
Next, we studied the significance of the second glycine residue at position 36 (G36) within the HPGG motif of EgΔ5D using the same approach as that for P34. Table 2 shows that the G36 residue within the HPGG motif could be substituted with several different amino acids without significantly impacting the Δ5 desaturase activity of EgΔ5D. The EgΔ5D-36S or EgΔ5D-36D mutant had about 100.8 or 99.2 % of Δ5 desaturase activity when compared to EgΔ5D, respectively.
The above functional studies at the P34 and G36 positions within the HPGG motif of EgΔ5D demonstrated that the HPGG motif could be changed without impacting the Δ5 desaturase activity. Specifically, the EgΔ5D-34G, EgΔ5D-36S and EgΔ5D-36D mutants were functionally equivalent to the wild type EgΔ5D. Comparison of the ARA in unesterified fatty acid (FFA), phospholipid (PL) and neutral lipid (NL) pools of Yarrowia transformants with EgΔ5D and EgΔ5D-34G showed that the P34G mutation did not affect the ARA distribution in these pools (Supplemental Fig. S2).
Improvement of Δ5 Desaturase Activity of EgΔ5D by Amino Acid Substitution Within the HDASH Motif
The HDASH motif was also claimed as one of the characteristics of Δ5 desaturases and necessary for its function to convert DGLA to ARA [34]. To test the hypothesis that the exact sequence of the HDASH motif (position 155–159) of EgΔ5D was required, we first selected the alanine residue at position 157 (A157) as a target. The Δ5 desaturase activity attributed to each mutation at A157 is summarized in Table 3. The data showed that almost all mutations at A157 greatly reduced the Δ5 desaturase activity of EgΔ5D. However, the EgΔ5D-157G and EgΔ5D-157S mutants retained about 96 and 94 % activity of wild type EgΔ5D, respectively.
We also studied the significance of the serine residue at position 158 (S158) within the HDASH motif of EgΔ5D. Table 4 shows that the S158 could be substituted with either an alanine or a glycine without substantially impacting the ∆5 desaturase activity of EgΔ5D. Specifically, the EgΔ5D-158A mutant had about 100.9 % activity of wild type EgΔ5D, while the EgΔ5D-158G mutation had about 107.7 % activity of wild type EgΔ5D. These data demonstrated that the HDASH motif could be changed without significantly reducing the Δ5 desaturase activity. In fact, the enzyme activity of EgΔ5D could be improved by substitution of the S158 within the HDASH motif with a glycine residue.
Both HPGG and HDASH Motifs were not Necessary in the Exact Form as Encoded for Δ5 Desaturase Activity of EgΔ5D
In order to study whether EgΔ5D could maintain strong Δ5 desaturase activity with mutations in both HPGG and HDASH motifs, we generated a series of double mutants. The results (Table 5) demonstrate that ∆5 desaturases could be constructed having variant HPGG and HDASH motifs that retain at least 64 % of ∆5 desaturase activity when compared to the wild type.
The proline residue within the HPGG motif can be substituted with glycine with simultaneous substitution of either (1) the alanine residue within the HDASH motif for glycine or (2) the serine residue within the HDASH motif for alanine or glycine. The proline residue within the HPGG motif can also be substituted with histidine with simultaneous substitution of the serine residue within the HDASH motif for either alanine or glycine. And, the second glycine residue within the HPGG motif can be substituted with serine with simultaneous substitution of the serine residue within the HDASH motif for either alanine or glycine. Specifically, the EgΔ5D-34G/157G, EgΔ5D-34G/158A and EgΔ5D-34H/158G double mutants had more than 80 % of the Δ5 desaturase activity of EgΔ5D, while EgΔ5D-34G/158G had about 97 % Δ5 desaturase activity of EgΔ5D. Further analyses showed that the ARA distribution in FFA, PL and NL pools of Yarrowia transformants with EgΔ5D-34G/158G was similar to Yarrowia transformants with EgΔ5D-34G or EgΔ5D (Supplemental Fig. S2), suggesting that the simultaneous substitutions (P34G and S158G) within HPGG and HDASH motifs did not change desaturase substrate specificity.
Increased Substrate Conversion of EgΔ5D-34G/158G with Double Mutations in HPGG and HDASH Motifs
In order to increase the substrate conversion of EgΔ5D-34G/158G, we optimized the codon usage of the N-terminal portion of the gene for expression in Y. lipolytica. The codon-optimized EgΔ5D-34G/158G, designated as “EgΔ5M”, had 48 bp changed in the first 204 bp of the coding region (23.5 %; Fig. 6), which resulted in optimization of 43 codons of the first 68 amino acids within the N-terminus of the protein (63.2 %). The amino acid sequence encoded by the codon-optimized EgΔ5M was identical to that of the EgΔ5D-34G/158G. EgΔ5M was used to replace the EgΔ5D of pDMW367-M4 to generate pDMW367-5M, containing a FBAIN::EgΔ5M::Pex20 chimeric gene.
We then studied the importance of the arginine or serine at position 347 that were found in the original clones of EgΔ5D. Based on EgΔ5M, the CGA codon for arginine at position 347 was changed to AGC codon to encode for serine, which was designated as EgΔ5M1. The synthetic EgΔ5M1 was used to replace the EgΔ5D of pDMW367-M4 to generate pDMW367-5M1, containing an FBAIN::EgΔ5M1::Pex20 chimeric gene.
The Δ5 desaturase activity of EgΔ5D, EgΔ5M and EgΔ5M1 is summarized in Table 6. GC analyses determined that there were about 3.6 % ARA and 10.8 % DGLA, 4.0 % ARA and 11.2 % DGLA, and 4.1 % ARA and 10.8 % DGLA of total fatty acids produced in the Yarrowia transformants with pDMW367-M4, pDMW367-5M, and pDMW367-5M1, respectively. It showed that the wild-type EgΔ5D converted about 24.8 % of DGLA to ARA; EgΔ5M converted 26.7 % of DGLA to ARA; and, EgΔ5M1 converted 27.6 % of DGLA to ARA. The fatty acid profile of Yarrowia transformants with EgΔ5M1 was almost identical to the profile of Yarrowia transformants with pDMW367-M4 as shown in Fig. 2c, except that more ARA was produced. These data demonstrated that the codon optimization of EgΔ5D improved its substrate conversion efficiency. Further, the amino acid at position 347 did affect the ∆5 desaturase activity of EgΔ5D, with a serine residue preferred over an arginine residue.
Discussion
Y. lipolytica has an established history of robust fermentation performance at commercial scale for processes including the production of food-grade citric acid for human consumption and single-cell protein for animal feeds [55]. Recently, Y. lipolytica has been used as a host for production of lipid-based compounds [14, 56, 57]. Some Y. lipolytica strains are oleaginous organisms that are able to accumulate up to 40 % dry cell weight as oil when starved for nitrogen in the presence of excess glucose as carbon source. However, LNA is the only PUFA that Y. lipolytica can synthesize de novo (Fig. 2a). Therefore, it is necessary to isolate genes encoding enzymes for every step of the “desaturation and elongation” pathways (Fig. 1) before genetically engineering Y. lipolytica to produce ARA, EPA and DHA oil. Δ5 desaturase is the enzyme responsible for the conversion of DGLA to ARA, and ETA to EPA. Although several Δ5 desaturase genes have been isolated from various organisms [26], more effective enzymes may help to improve the production of commercially important LC-PUFA.
Previous studies have indicated that Euglena was able to synthesize ARA, EPA and DHA through the “Δ9 elongase/Δ8 desaturase” pathway [16, 38, 45]. In this report, the gene encoding a Δ5 desaturase from E. gracilis was isolated and characterized. Our results indicated two nucleotide sequences with a difference of two base pairs that would result in either arginine or serine at position 347. This discrepancy was most likely generated from PCR amplification or during cDNA generation. BlastP searches showed that the amino acid sequence of EgΔ5D shares <40 % identity with any Δ5 desaturase found in Genbank; PtΔ5D [47] was the most similar one (about 39 %), suggesting that the primary structure of EgΔD5 is quite different from those Δ5 desaturase genes previously isolated. Amino acid sequence alignment also shows that EgΔ5D has about 20 % identity with EgΔ8D [16, 38] and about 25.5 % identity with EgΔ4D [45]. These data suggest that EgΔ5D is evolutionary closer to Δ5 desaturase than the Δ4 or Δ8 desaturases. Functional analyses of EgΔ5D in Y. lipolytica strains Y4036U and Y2224 revealed that it has strong Δ5 desaturase activity, with more than 24 % substrate conversion of DGLA to ARA, and it is not a Δ5/Δ6 bifunctional enzyme.
The HPGG motif is expected to be on the cytochrome b 5 surface, in contact with the heme through van der Waals interactions [27]. The conserved HPGG motif was thought to be essential in maintaining cytochrome b 5 electron transfer function, with the histidine serving as a heme axial ligand [50]. Substitution of the histidine residue of the HPGG motif with alanine in the Δ6 desaturase cytochrome b 5 domain of starflower, rat and algae abolished Δ6 desaturase activity [51–53]. It is expected that H33 of EgΔ5D should also be essential for its function.
The HPGG motif itself has a unique structure. The proline residue of the HPGG stretch is located in a turn between two consecutive helices (Fig. 5) and was thought to be important in protein folding and in maintaining cytochrome b 5 protein stability [32]. The three-carbon side chain of proline is bonded to both the nitrogen and the carbon of the peptide backbone to form a five-member ring that greatly restricted its conformational freedom. The nonpolar characteristic of this ring structure may create a hydrophobic spot within the HPGG motif. On the other hand, the glycine possesses the smallest amino acid side chain, hydrogen, which can allow for greater flexibility in local structure. It is likely that the combination of proline and glycine residues within the cytochrome b 5 HPGG motif is an important factor affecting both the structural position of the hydrophobic heme pocket and the appropriate orientation of the heme group within the heme pocket relative to the desaturase catalytic site. Surprisingly, our results indicate that the proline residue of the HPGG motif is not essential for electron transfer from the heme group of the cytochrome b 5 domain to the catalytic diiron cluster of EgΔ5D. Most substitution mutants at P34 displayed at least 70 % of the wild type EgΔ5D activity. The EgΔ5D-34G (HgGG) mutant had greater than 98 % of the wild type EgΔ5D activity, demonstrating that the proline residue of HPGG motif is not required for the enzyme activity of EgΔ5D (Table 1).
It is noteworthy that aspartate substitutions in EgΔ5D-34D and EgΔ5D-36D exhibit different effects on desaturase activity. Compared to free cytochrome b 5, while there are several conserved acidic amino acids, there is a characteristic reduction in the number of aspartate and glutamate residues in the vicinity of the HPGG motif of cytochrome b 5 domain of desaturases. This reduced number of acidic residues around the heme pocket is thought to contribute to stabilizing nonpolar intermolecular interactions between the cytochrome b 5 and desaturase domains [27, 54]. Substitutions involving aspartate or glutamate residues in the HPGG motif may affect the interface geometry of electron donor/acceptor docking that is exhibited in desaturase activity due to altered electron transfer. We also found that substitutions for G36 of EgΔ5D resulted in mutants with strong Δ5 desaturase activity. The most functional mutants were the small, slightly polar serine replacement, EgΔ5D-36S, and the acidic substitution with aspartate, EgΔ5D-36D. The activities of these two mutants are about the same as the wild type EgΔ5D (Table 2).
The amino acid sequence of the first His-rich motif, HX(3, 4)H, of EgΔ5D is HDASH located from residues 155 to 159. The HDASH motif has been suggested as one of the characteristics of Δ5 desaturases and necessary for its function to convert DGLA to ARA in any transformed organisms [34]. Sequence analyses showed that there are natural variants of the HDASH motif in Δ5 desaturases, for example, PiΔ5D [46] has the sequence of HDsSH, the Δ5 desaturase from Thraustochytrium sp. ATCC 21685 (GenBank accession #: AAM09687) has the sequence of HemgH, the Δ5 desaturase from Leishmania major strain Friedlin (GenBank accession #: CAJ07076) has the sequence of HeAgH, the Δ5 desaturase from Atlantic salmon (GenBank accession #: AAL82631) has the sequence of HDygH, and PtΔ5D [47] has the sequence of HDAnH in the corresponding location. This suggests that the HDASH motif is not an invariant characteristic of Δ5 desaturases, and may be not required for Δ5 desaturase activity. We suggest that the two His residues of HDASH motif participate in the coordination of the diiron center (Fig. 5), but the other three residues (DAS) residues between the two His residues can be modified.
Systemic substitution studies (Tables 3, 4) at positions A157 and S158 within the HDASH motif of EgΔ5D demonstrated that these two residues could be replaced, and the mutants retained good Δ5 desaturase activity. The EgΔ5D-157G and EgΔ5D-157S mutants had about 96 and 94 % of the wild type EgΔ5D activity, respectively. Since PiΔ5D has an HDsSH motif [46], it is not surprising that EgΔ5D-157S with sequence HDsSH functioned well in Yarrowia. We also found that S158 could be substituted with either alanine or glycine. The alanine, glycine and serine are interchangeable within the HDASH motif of EgΔ5S; furthermore, the enzyme activity of EgΔ5D could be improved with a motif of HDAgH instead of HDASH (Table 4).
To determine whether at least one motif, HPGG or HDASH, is required for the enzyme activity of EgΔ5D, a series of mutants with mutations in both the HPGG and HDASH motifs was generated (Table 5). Some double mutants such as EgΔ5D-34G/157G, EgΔ5D-34G/158A and EgΔ5D-34H/158G had more than 80 % of wild type EgΔ5D activity, while EgΔ5D-34G/158G had almost the same activity as wild type EgΔ5D. Therefore, neither the HPGG nor the HDASH motif is necessary in the exact form as encoded for the activity of EgΔ5D.
Distribution analyses (Supplemental Fig. S2) of ARA in FFA, PL and NL pools of Yarrowia transformants with EgΔ5D shows that more ARA loaded in PL pool than that in FFA pool, suggesting that EgΔ5D is also an acyl-lipid desaturase, just like other front-end desaturases from lower plants, fungi and algae [26]. ARA distribution comparison of Yarrowia transformants with EgΔ5D-34G and EgΔ5D-34G/158G with wild type EgΔ5D shows that either single mutation within HPGG motif (P34G), or simultaneous mutations within HPGG (P34G) and HDASH (S158G) motifs does not change its fatty acid distribution pattern (Supplemental Fig. S2), and therefore these changes in EgΔ5D do not affect its substrate specificity.
An effective Δ5 desaturase is required for efficient conversion of DGLA to ARA or ETA to EPA (Fig. 1) in engineered Y. lipolytica or other organisms to produce commercially valuable LC-PUFA. We employed two approaches to improve the enzyme activity of the double mutant EgΔ5D-34G/158G. The optimization (Fig. 6) of the 43 codons of the 68 amino acids within the N-terminal portion of EgΔ5D-34G/158G, EgΔ5M, improved substrate conversion (Table 6). This improvement may relate to the rate of translation. Recent reports suggest that sequence at the beginning of a gene can influence translation, and the mRNA structure at the 5′ end of an mRNA can affect protein levels [58, 59]. Next, we substituted the arginine residue at position 347 with the serine which was identified in our original PCR products. Surprisingly, this R347S substitution in codon optimized EgΔ5M1 further improved substrate conversion (Table 6). These data suggest that some un-conserved amino acids among different Δ5 desaturases may be good targets for protein evolution to generate improved enzymes. At this stage, the improved EgΔ5D, both EgΔ5M and EgΔ5M1 should enable us to engineer Yarrowia and other organisms to produce high levels of ARA, EPA and DHA.
In conclusion, our studies suggest that the exact sequences of the HPGG and HDASH motifs are not necessary for the function of EgΔ5D. Several amino acids within these two motifs can be changed individually, or simultaneously, without significantly reducing the enzyme activity or altering its substrate specificity. In some cases such as EgΔ5D-36D, EgΔ5D-36S, and EgΔ5D-158G mutants, the Δ5 desaturase activity can be improved. In order to fully understand the function of EgΔ5D, the roles of the HX(2, 3)HH and (H/Q)X(2, 3)HH motifs as well as the un-conserved amino acids need to be studied in the future.
Abbreviations
- ALA:
-
Alpha-linolenic acid (ALA 18:3n-3)
- ARA:
-
Arachidonic acid (C20:4n-6)
- ATCC:
-
American Type Culture Collection (Rockville, MD)
- DAG:
-
Diacylglycerol
- DdΔ5D:
-
Δ5 Desaturase of Dictyostelium discoideum
- DGLA:
-
Dihomo-gamma-linoleic acid (C20:3n-6)
- DHA:
-
Docosahexaenoic acid (C22:6n-3)
- DPA:
-
Docosapentaenoic acid (22:5n-3)
- EgΔ4D:
-
Δ4 Desaturase of Euglena gracilis
- EgΔ5D:
-
Δ5 Desaturase of Euglena gracilis
- EgΔ8D:
-
Δ8 Desaturase of Euglena gracilis
- EG Media:
-
Euglena growth media
- FFA:
-
Unesterified fatty acids
- FOA:
-
5-Fluoro-orotic acid FOA
- EPA:
-
Eicosapentaenoic acid (C20:5n-3)
- ETA:
-
Eicosatetraenoic acid (C20:4n-3)
- GC:
-
Gas chromatography
- HGM:
-
High glucose media
- His-rich motif:
-
Histidine-rich motif
- IgΔ5D:
-
Δ5 Desaturase of Isochrysis galbana
- LC-PUFA:
-
Long chain polyunsaturated fatty acids
- LNA:
-
Linoleic acid (18:2n-6)
- MaΔ5D:
-
Δ5 Desaturase of Mortierella alpina
- MMLeu:
-
Minimal media + leucine
- NL:
-
Neutral lipids
- OtΔ5D:
-
Δ5 Desaturase of Ostreococcus tauri
- PCR:
-
Polymerase chain reaction
- PiΔ5D:
-
Δ5 Desaturase of Pythium irregulare
- PL:
-
Phospholipids
- PlΔ8D:
-
Δ8 Desaturase of Pavlova lutheri
- PmΔ5D:
-
Δ5 Desaturase of Phytophthora megasperma
- PtΔ5D:
-
Δ5 Desaturase of Phaeodactylum tricornutum
- PUFA:
-
Polyunsaturated fatty acids
- RACE:
-
Rapid amplification of cDNA ends
- TAG:
-
Triacylglycerol
- TaΔ5D:
-
Δ5 Desaturase of Thraustochytrium aureum
- TsΔ8D:
-
Δ8-Sphingolipid desaturase of Thalassiosira pseudonana
- α:
-
Alpha
- γ:
-
Gamma
- Δ:
-
Delta
- #:
-
Number
References
Riediger ND, Othman RA, Suh M, Moghadasian MH (2009) A systemic review of the roles of n-3 fatty acids in health and disease. J Am Diet Assoc 109:668–679
Freeman LM (2010) Beneficial effects of omega-3 fatty acids in cardiovascular disease. J Small Anim Pract 51:462–470
Mourek J, Mourek J Jr (2011) Developmentally dependent and different roles of fatty acids OMEGA-6 and OMEGA-3. Prague Med Rep 112:81–92
Mayurasakorn K, Williams JJ, Ten VS, Deckelbaum RJ (2011) Docosahexaenoic acid: brain accretion and roles in neuroprotection after brain hypoxia and ischemia. Curr Opin Clin Nutr Metab Care 14:158–167
Uddin M, Levy BD (2011) Resolvins: natural agonists for resolution of pulmonary inflammation. Prog Lipid Res 50:75–88
Serhan CN (2007) Resolution phase of inflammation: novel endogenous anti-inflammatory and proresolving lipid mediators and pathways. Annu Rev Immunol 25:101–137
Kiso Y (2011) Pharmacology in health foods: effects of arachidonic acid and docosahexaenoic acid on the age-related decline in brain and cardiovascular system function. J Pharmacol Sci 115:471–475
Weitz D, Weintraub H, Fisher E, Schwartzbard AZ (2010) Fish oil for the treatment of cardiovascular disease. Cardiol Rev 18:258–263
Yokoyama M, Origasa H, Matsuzaki M, Matsuzawa Y, Saito Y, Ishikawa Y, Oikawa S, Sasaki J, Hishida H, Itakura H, Kita T, Kitabatake A, Nakaya N, Sakata T, Shimada K, Shirato K (2007) Effects of eicosapentaenoic acid on major coronary events in hypercholesterolaemic patients (JELIS): a randomised open-label, blinded endpoint analysis. Lancet 369:1090–1098
Féart C, Peuchant E, Letenneur L, Samieri C, Montagnier D, Fourrier-Reglat A, Barberger-Gateau P (2008) Plasma eicosapentaenoic acid is inversely associated with severity of depressive symptomatology in the elderly: data from the Bordeaux sample of the three-city study. Am J Clin Nutr 87:1156–1162
Costa LG (2007) Contaminants in fish: risk-benefit considerations. Arh Hig Rada Toksikol 58:567–574
Streekstra H (2010) Arachidonic acid: fermentative production by Mortierella fungi. In: Cohen Z, Ratledge C (eds) Single cell oils, 2nd edn. AOCS Press, Urbana
Barclay W, Weaver C, Metz J, Hansen J (2010) Development of a docosahexaenoic acid production technology using Schizochytrium: historical perspective and update. In: Cohen Z, Ratledge C (eds) Single cell oils, 2nd edn. AOCS Press, Urbana
Zhu Q, Xue Z, Yadav N, Damude H, Pollak D, Rupert R, Seip J, Hollerbach D, Macool D, Zhang H (2010) Metabolic engineering of an oleaginous yeast for the production of omega-3 fatty acids. In: Cohen Z, Ratledge C (eds) Single cell oils, 2nd edn. AOCS Press, Urbana
Metz JG, Roessler P, Facciotti D, Levering C, Dittrich F, Lassner M, Valentine R, Lardizabal K, Domergue F, Yamada A, Yazawa K, Knauf V, Browse J (2001) Production of polyunsaturated fatty acids by polyketide synthases in both prokaryotes and eukaryotes. Science 293:290–293
Wallis JG, Browse J (1999) The Δ8-desaturase of Euglena gracilis: an alternate pathway for synthesis of 20-carbon polyunsaturated fatty acids. Arch Biochem Biophys 365:307–316
Pereira SL, Leonard AE, Mukerji P (2003) Recent advances in the study of fatty acid desaturases from animals and lower eukaryotes. Prostaglandins Leukot Essent Fatty Acids 68:97–106
Qiu X, Hong H, MacKenzie SL (2001) Identification of a delta 4 fatty acid desaturase from Thraustochytrium sp. involved in the biosynthesis of docosahexanoic acid by heterologous expression in Saccharomyces cerevisiae and Brassica juncea. J Biol Chem 276:31561–31566
Qi B, Fraser T, Mugford S, Dobson G, Sayanova O, Butler JJ, Napier A, Stobart AK, Lazarus CM (2004) Production of very long chain polysaturated omega-3 and omega-6 fatty acids in plants. Nat Biotechnol 22:739–745
Kajikawa M, Yamato KT, Kohzu Y, Nojiri M, Sakuradani E, Shimizu S, Sakai Y, Fukuzawa H, Ohyama K (2004) Isolation and characterization of delta(6)-desaturase, an ELO-like enzyme and delta(5)-desaturase from the liverwort Marchantia polymorpha and production of arachidonic and eicosapentaenoic acids in the methylotrophic yeast Pichia pastoris. Plant Mol Biol 54:335–352
Damude HG, Kinney AJ (2008) Engineering oilseeds to produce nutritional fatty acids. Physiol Plant 132:1–10
Venegas-Calerón M, Sayanova O, Napier JA (2010) An alternative to fish oils: metabolic engineering of oil-seed crops to produce omega-3 long chain polyunsaturated fatty acids. Prog Lipid Res 49:108–119
Tavares S, Grotkjær T, Obsen T, Haslam RP, Napier JA, Gunnarsson N (2011) Metabolic engineering of Saccharomyces cerevisiae for production of eicosapentaenoic acid, using a novel {delta}5-desaturase from Paramecium tetraurelia. Appl Environ Microbiol 77:1854–1861
Michaelson LV, Lazarus CM, Griffiths G, Napier JA, Stobart AK (1998) Isolation of a delta5-fatty acid desaturase gene from Mortierella alpina. J Biol Chem 273:19055–19059
Knutzon DS, Thurmond JM, Huang YS, Chaudhary S, Bobik EG Jr, Chan GM, Kirchner SJ, Mukerji P (1998) Identification of delta5-desaturase from Mortierella alpina by heterologous expression in bakers’ yeast and canola. J Biol Chem 273:29360–29366
Meesapyodsuk D, Qiu X (2012) The front-end desaturase: structure, function, evolution and biotechnological use. Lipids 47:227–237
Sperling P, Heinz E (2001) Desaturases fused to their electron donor. Eur J Lipid Sci Technol 103:158–180
Napier JA, Michaelson LV, Sayanova O (2003) The role of cytochrome b 5 fusion desaturases in the synthesis of polyunsaturated fatty acids. Prostaglandins Leukot Essent Fatty Acids 68:135–143
Schenkman JB, Jansson I (2003) The many roles of cytochrome b5. Pharmacol Ther 97:139–152
Stukey JE, McDonough VM, Martin CE (1990) The OLE1 gene of Saccharomyces cerevisiae encodes the delta 9 fatty acid desaturase and can be functionally replaced by the rat stearoyl-CoA desaturase gene. J Biol Chem 265:20144–20149
Shanklin J, Guy JE, Mishra G, Lindqvist Y (2009) Desaturases: emerging models for understanding functional diversification of diiron-containing enzymes. J Biol Chem 284:18559–18563
Lin YW, Ying TL, Liao LF (2009) Dynamic consequences of mutating the typical HPGG motif of apocytochrome b 5 revealed by computer simulation. Chin Chem Let 200:631–634
Shanklin J, Whittle E, Fox BG (1994) Eight histidine residues are catalytically essential in a membrane-associated iron enzyme, stearoyl-CoA desaturase, and are conserved in alkane hydroxylase and xylene monooxygenase. Biochemistry 33:12787–12794
Knutzon D, Mukerji P, Huang YS, Thurmond J, Chaudhary S (1999) Methods and compositions for synthesis of long chain poly-unsaturated fatty acids. US Patent 5,972,664
Yadav NS, Zhu Q, Zhang H (2009) Δ12 desaturases suitable for altering levels of polyunsaturated fatty acids in oleaginous yeast. US Patent 7,504,259
Macool, DJ, Xue Z, Zhu Q (2008) A Mortierella alpina c16/18 fatty acid elongase. US Patent 7,470,532
Damude HG, McGonigle B, Zhu Q, Xue Z (2011) Delta-9 elongases and their use in making polyunsaturated fatty acids. US Patent 8,049,062
Damude HG, He H, Liao D-I, Zhu Q (2011) Mutant delta-8 desaturase genes engineered by targeted mutagenesis and their use in making polyunsaturated fatty acids. US Patent 8,026,089
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor
Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (2010) Current protocols in molecular biology. Wiley, New York
Fournier P, Abbas A, Chasles M, Kudla B, Ogrydziak D, Yaver D, Xuan JW, Peito A, Ribe AM, He F, Gaillardin CC (1993) Colocalization of centromeric and replicative functions on autonomously replicating sequences isolated from the yeast Yarrowia lipolytica. Proc Natl Acad Sci USA 90:4912–4916
Hong S-Y, Seip J, Walters-Pollak D, Rupert R, Jackson R, Xue Z, Zhu Q (2011) Engineering Yarrowia lipolytica to express secretory invertase with strong FBA1IN promoter. Yeast 29:59–72
Chen DC, Beckerich JM, Gaillardin C (1997) One-step transformation of the dimorphic yeast Yarrowia lipolytica. Appl Microbiol Biotechnol 48:232–235
Cahoon EB, Ripp KG, Hall SE, Kinney AL (2001) Formation of conjugated Δ8, Δ10-double bonds by Δ12-oleic acid desaturase-related enzymes: biosynthetic origin of calendic acid. J Biol Chem 276:2637–2643
Meyer A, Cirpus P, Ott C, Schlecker R, Zahringer U, Heinz E (2003) Biosynthesis of docosahexaenoic acid in Euglena gracilis: biochemical and molecular evidence for the involvement of a delta4-fatty acyl group desaturase. J Biochem 42:9779–9788
Hong H, Datla N, MacKenzie SL, Qiu X (2002) Isolation and characterization of a Δ5 FA desaturase from Pythium irregulare by heterologous expression in Saccharomyces cerevisiae and oilseed crops. Lipids 37:863–868
Domergue F, Lerchl J, Zahringer U, Heinz E (2002) Cloning and functional characterization of Phaeodactylum tricornutum front-end desaturases involved in eicosapentaenoic acid biosynthesis. Eur J Biochem 269:4105–4113
Damude H, Zhu Q (2007) Delta-8 desaturase and its use in making polyunsaturated fatty acids. US Patent 7,943,823
Kobayashi T, Sakaguchi K, Matsuda T, Abe E, Hama Y, Hayashi M, Honda D, Okita Y, Sugimoto S, Okino N, Ito M (2011) Increase of eicosapentaenoic acid in thraustochytrids through thraustochytrid ubiquitin promoter-driven expression of a fatty acid {delta}5 desaturase gene. Appl Environ Microbiol 77:3870–3876
Lederer F (1994) The cytochrome b4-fold: an adaptable module. Biochimie 76:674–692
Sayanova O, Shewry PR, Napier JA (1999) Histidine-41 of the cytochrome b5 domain of the borage delta6 fatty acid desaturase is essential for enzyme activity. Plant Physiol 121:641–646
Guillou H, D’Andrea S, Rioux V, Barnouin R, Dalaine S, Pedrono F, Jan S, Legrand P (2004) Distinct roles of endoplasmic reticulum cytochrome b5 and fused cytochrome b5-like domain for rat Δ6-desaturase activity. J Lipid Res 45:32–40
Hongsthong A, Subudhi S, Sirijuntarut M, Kurdrid P, Cheevadhanarak S, Tanticharoen M (2006) Revealing the complementation of ferredoxin by cytochrome b5 in the Spirulina-Δ6-desaturation reaction by N-terminal fusion and co-expression of the fungal-cytochrome b5 domain and Spirulina-Δ6-acyl-lipid desaturase. Appl Microbiol Biotechnol 72:1192–1201
Gostincar C, Turk M, Gunde-Cimerman N (2010) The evolution of fatty acid desaturases and cytochrome b5 in eukaryotes. J Membr Biol 233:63–72
Ratledge C (2010) Single cell oils for the 21st century. In: Cohen Z, Ratledge C (eds) Single cell oils, 2nd edn. AOCS Press, Urbana
Beopoulos A, Nicaud JM, Gaillardin C (2011) An overview of lipid metabolism in yeasts and its impact on biotechnological processes. Appl Microbiol Biotechnol 90:1193–1206
Sabirova JS, Haddouche R, Van Bogaert IN, Mulaa F, Verstraete W, Timmis KN, Schmidt-Dannert C, Nicaud JM, Soetaert W (2011) The ‘LipoYeasts’ project: using the oleaginous yeast Yarrowia lipolytica in combination with specific bacterial genes for the bioconversion of lipids, fats and oils into high-value products. Microb Biotechnol 4:47–54
Kudla G, Murray AW, Tollervey D, Plotkin JB (2009) Coding sequence determinants of gene expression in Escherichia coli. Science 324:255–258
Kahali B, Ahmad S, Ghosh TC (2011) Selective constraints in yeast genes with differential expressivity: codon pair usage and mRNA stability perspectives. Gene 481:76–82
Acknowledgments
We are grateful to Ethel Jackson and Henry Bryndza for their strong support. We thank every member of our Omega-3 team for their suggestions and discussions, Raymond Jackson and the DNA sequencing lab for their technical service, and Kelley Norton and Arthur Kruckeberg for critical reading of this manuscript.
Conflict of interest
There are no actual or potential conflicts of interest.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Pollak, D.W., Bostick, M.W., Yoon, H. et al. Isolation of a Δ5 Desaturase Gene from Euglena gracilis and Functional Dissection of Its HPGG and HDASH Motifs. Lipids 47, 913–926 (2012). https://doi.org/10.1007/s11745-012-3690-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11745-012-3690-1