Abstract
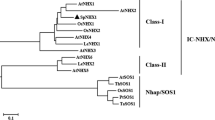
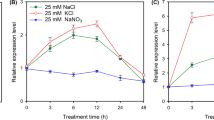
Growth and development of plants are greatly affected by salinity. The salt overly sensitive (SOS) pathway plays a vital role in plants responding to salt stress, and has been reported to contain three components: SOS3 (calcineurin B-like protein 4, CBL4), SOS2 (CBL-interacting protein kinase 24, CIPK24) and SOS1. Our previous study demonstrated that transgenic yeast and Arabidopsis could tolerate salt better when expressed with the cell membrane Na+/H+ antiporter SOS1 from the halophyte Sesuvium portulacastrum. Here, a new CIPK gene (SpCIPK8) and CBL gene (SpCBL10) have been isolated from S. portulacastrum. The expression of SpCIPK8 and SpCBL10 was induced by salinity in roots of S. portulacastrum. An interaction between SpCBL10 and SpCIPK8 was demonstrated in yeast two-hybrid assays. Subsequent analysis found that SpCBL10 could bind the C-terminus of SpCIPK8. Yeast co-expressing SpSOS1, SpCIPK8 and SpCBL10 genes grew better and accumulated more potassium (K+) and less sodium (Na+) under salt stress than yeast that expressed only one or two of these genes, indicating that Na+ was excluded from the cells. Furthermore, we found that the SpCBL10/SpCIPK8 complex regulates the cell membrane Na+/H+ antiporter SpSOS1 to enhance yeast salt tolerance by binding the two serine residues at amino acid positions 1144 and 1146 in the conserved DSPS motif at the C-terminus of SpSOS1. Future studies of the SOS pathway will be greatly aided by these results, which suggest some candidate genes for improving plant salt tolerance.






Similar content being viewed by others
Data availability
We ensured that all datasets were presented in the main manuscript or additional supporting files.
References
Arabbeigi M, Arzani A, Majidi MM et al (2018) Expression pattern of salt tolerance-related genes in Aegilops cylindrica. Physiol Mol Biol Plants 24(1):61–73
Arzani A, Ashraf M (2016) Smart engineering of genetic resources for enhanced salinity tolerance in crop plants. Crit Rev Plant Sci 35(3):146–189
Boudsocq M, Laurie’re C (2005) Osmotic signaling in plants: multiple pathways mediated by emerging kinase families. Plant Physiol 138:1185–1194
Cheong YH, Pandey GK, Grant JJ et al (2007) Two calcineurin B-like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis. Plant J 52:223–239
Fan Y, Yin X, Xie Q et al (2019) Co-expression of SpSOS1 and SpAHA1 in transgenic Arabidopsis plants improves salinity tolerance. BMC Plant Biol 19:74
Feki K, Quintero FJ, Pardo JM et al (2011) Regulation of durum wheat Na+/H+ exchanger TdSOS1 by phosphorylation. Plant Mol Biol 76:545–556
Flowers TJ, Clomer TD (2008) Salinity tolerance in halophytes. New Phytol 179:945–963
Flowers TJ, Galal H, Bromham L (2010) Evolution of halophytes: multiple origins of salt tolerance in land plants. Funct Plant Biol 37:604–612
Ghnaya T, Slama I, Messedi D et al (2007) Cd-induced growth reduction in the halophyte Sesuvium portulacastrum is significantly improved by NaCl. J Plant Res 120:309–316
Gong D, Guo Y, Jagendorf AT et al (2002) Biochemical characterization of the Arabidopsis protein kinase SOS2 that functions in salt tolerance. Plant Physiol 131:256–264
Guo Y, Halfter U, Ishitani M et al (2001) Molecular characterization of functional domains in the protein kinase SOS2 that is required for salt tolerance. Plant Cell 13:1383–1399
Guo Y, Qiu QS, Quintero FJ et al (2004) Transgenic evaluation of activated mutant alleles of SOS2 reveals a critical requirement for its kinase activity and C-terminal regulatory domain for salt tolerance in Arabidopsis thaliana. Plant Cell 16:435–449
Hashimoto K, Eckert C, Anschütz U et al (2012) Phosphorylation of calcineurin B-like (CBL) calcium sensor proteins by their CBL-interacting protein kinases (CIPKs) is required for full activity of CBL-CIPK complexes toward their target proteins. J Biol Chem 287:7956–7968
Ho CH, Lin SH, Hu HC et al (2009) CHL1 functions as a nitrate sensor in plants. Cell 138:1184–1194
Kiani R, Arzani A, Maibody SAMM et al (2021) Morpho-physiological and gene expression responses of wheat by Aegilops cylindrica amphidiploids to salt stress. Plant Cell Tissue Org Culture 144:619–639
Knighton DR, Zheng JH, Ten Eyck LF et al (1991) Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 253:407–414
Kolukisaoglu U, Weinl S, Blazevic D et al (2004) Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol 134:43–58
Kronzucker HJ, Britto DT (2011) Sodium transport in plants: a critical review. New Phytol 189:54–81
Léran S, Edel KH, Pervent M et al (2015) Nitrate sensing and uptake in Arabidopsis are enhanced by ABI2, a phosphatase inactivated by the stress hormone abscisic acid. Sci Signal 8(375):43
Li L, Kim BG, Cheong YH et al (2006) A Ca2+ signaling pathway regulates a K+ channel for low-K response in Arabidopsis. Proc Natl Acad Sci USA 103:12625–12630
Liu J, Zhu JK (1998) A calcium sensor homolog required for plant salt tolerance. Science 280:1943–1945
Liu J, Ishitani M, Halfter U et al (2000) The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc Natl Acad Sci USA 97:3730–3734
Luan S (2009) The CBL-CIPK network in plant calcium signaling. Trends Plant Sci 14:37–42
Ma DM, Xu WR, Li HW et al (2014) Co-expression of the Arabidopsis SOS genes enhances salt tolerance in transgenic tall fescue (Festuca arundinacea Schreb.). Protoplasma 251:219–231
Mahi HE, Perez-Hormaeche J, De Luca A et al (2019) A critical role of sodium flux via the plasma membrane Na+/H+ exchanger SOS1 in the salt tolerance of rice. Plant Physiol 180:1046–1065
Mao J, Manik SM, Shi S et al (2016) Mechanisms and physiological roles of the CBL-CIPK networking system in Arabidopsis thaliana. Genes (basel) 7:62
Messedi D, Sleimi N, Abdelly C (2001) Salt tolerance in Sesuvium portulacastrum. Plant Nutri 92:406–407
Munns R (2002) Comparative physiology of salt and water stress. Plant Cell Environ 25:239–250
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 58:651–681
Olías R, Eljakaoui Z, Li J et al (2009) The plasma membrane Na+/H+ antiporter SOS1 is essential for salt tolerance in tomato and affects the partitioning of Na+ between plant organs. Plant Cell Environ 32:904–916
Park HJ, Kim WY, Yun DJ (2016) A new insight of salt stress signaling in plant. Mol Cells 39:447–459
Qiu QS, Guo Y, Dietrich MA et al (2002) Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci USA 99:8436–8441
Quan R, Lin H, Mendoza I et al (2007) SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 19:1415–1431
Quintero FJ, Martinez-Atienza J, Villalta I et al (2011) Activation of the plasma membrane Na/H antiporter Salt-Overly-Sensitive 1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain. Proc Natl Acad Sci USA 108:2611–2616
Ragel P, Ródenas R, García-Martín E et al (2015) The CBL-interacting protein kinase CIPK23 regulates HAK5-mediated high-affinity K+ uptake in Arabidopsis roots. Plant Physiol 169:2863–2873
Sanchez-Barrena MJ, Martinez-Ripoll M, Albert A (2013) Structural biology of a major signaling network that regulates plant abiotic stress: the CBL-CIPK mediated pathway. Int J Mol Sci 14:5734–5749
Scotto-Lavino E, Du GW, Frohman MA (2006a) 5′ end cDNA amplification using classic RACE. Nat Protoc 1:2555–2562
Scotto-Lavino E, Du GW, Frohman MA (2006b) 3’ end cDNA amplification using classic RACE. Nat Protoc 1:2742–2745
Snedden WA, Fromm H (1998) Calmodulin, calmodulin-related proteins and plant responses to the environment. Trends Plant Sci 3:299–304
Song A, Lu J, Jiang J et al (2012) Isolation and characterisation of Chrysanthemum crassum SOS1, encoding a putative plasma membrane Na(+)/H(+) antiporter. Plant Biol (stuttg) 14:706–713
Tang RJ, Zhao FG, Garcia VJ et al (2015) Tonoplast CBL-CIPK calcium signaling network regulates magnesium homeostasis in Arabidopsis. Proc Natl Acad Sci USA 112:3134–3139
Tang RJ, Liu H, Bao Y et al (2010) The woody plant poplar has a functionally conserved salt overly sensitive pathway in response to salinity stress. Plant Mol Biol 74:367–380
Tester M, Davenport R (2003) Na+ tolerance and Na+ transport in higher plants. Ann Bot 91:503–527
Weinl S, Kudla J (2009) The CBL-CIPK Ca2+-decoding signaling network: function and perspectives. New Phytol 184:517–528
Wu SJ, Ding L, Zhu JK (1996) SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell 8:617–627
Yang CL, Zhou Y, Fan J et al (2015) SpBADH of the halophyte Sesuvium portulacastum strongly confers drought tolerance through ROS scavenging in transgenic Arabidopsis. Plant Physiol Biochem 96:377–387
Yin XX, Xia YQ, Xie Q et al (2020) The protein kinase complex CBL10-CIPK8-SOS1 functions in Arabidopsis to regulate salt tolerance. J Exp Bot 71:1801–1814
Zhang WD, Wang P, Bao Z et al (2017) SOS1, HKT1;5, and NHX1 synergistically modulate Na+ homeostasis in the halophytic grass Puccinellia tenuiflora. Front Plant Sci 8:576
Zhang TT, Cui Z, Li YX et al (2021) Genome-wide identification and expression analysis of MYB transcription factor superfamily in Dendrobium catenatum. Front Genet 12:714696
Zhou Y, Yin X, Duan R et al (2015) SpAHA1 and SpSOS1 coordinate in transgenic yeast to improve salt tolerance. PLoS ONE 10:e0137447
Zhou Y, Lai Z, Yin X et al (2016) Hyperactive mutant of a wheat plasma membrane Na+/H+ antiporter improves the growth and salt tolerance of transgenic tobacco. Plant Sci 253:176–186
Acknowledgements
This research was supported by the National Key Research and Development Program of China (2018YFE0207203-2), the Education Department of Hainan Province (Hnky2021-19), Hainan Provincial Natural Science Foundation of China (318QN189), the National Natural Science Foundation of China (31660253), and the Startup funding from Hainan University (KYQD(ZR)1845).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by S. Renault.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11738_2023_3518_MOESM1_ESM.doc
Table S1 Primers sequences used in this study. Indicated are information about the primer names and primer sequences. Restriction sites are underlined. Table S2 Percentage of homology between SpCBL10 and Arabidopsis CBL10 proteins. Table S3. Percentage of homology between SpCIPK8 and Arabidopsis CIPK proteins (DOC 84 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhou, Y., Zhu, Y., Li, W. et al. Heterologous expression of Sesuvium portulacastrum SOS-related genes confer salt tolerance in yeast. Acta Physiol Plant 45, 58 (2023). https://doi.org/10.1007/s11738-023-03518-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11738-023-03518-7