Abstract
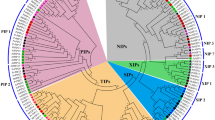
Plasma membrane H+-ATPase, which generates the proton gradient across the outer membrane of plant cells, plays a crucial role in the regulation of many physiological processes. In several species, PM H+-ATPase is encoded by a multigene family (HA). In contrast to Arabidopsis thaliana, Nicotiana plumbaginifolia and Oryza sativa, there is little information about the HA family in cucumber. Thus, in this study the first comprehensive analysis of the Cucumis sativus HA family was made. Using Blastn searches of cucumber whole genome shotgun reads from the GenBank database with 12 A. thaliana cDNAs encoding AtAHA1-12 proteins as the query, we retrieved a total of ten sequences, which significantly matched with AtAHAs. Phylogenetic analysis of CsHAs based on protein alignment revealed the representatives of three (I, II and IV) among five subfamilies known in other plant species. Expression patterns of genes showed their specific profiles during different phases of cucumber development and under specific exogenous factors. We also identified the putative cis-acting elements in CsHA promoters. Furthermore, the links between the patterns of cis-regulatory elements and tissue-specific or environmental-responding expression of genes as a first step to understanding the regulatory mechanism of HA in cucumber was performed. Our results provide a valuable basis for future studies aimed at explaining the role of particular H+-ATPase isoforms in cucumber responsiveness to the specific environmental factors.





Similar content being viewed by others
Abbreviations
- ABA:
-
Abscisic acid
- ACC:
-
1-Aminocyclopropane-1-carboxylic acid
- HA:
-
H+-ATPase
- CsHA:
-
Cucumis sativus H+-ATPase
- CRE:
-
Cis regulatory element
- ESTs:
-
Expressed sequence tags
- GA3 :
-
Gibberellic acid
- IAA:
-
Indole-3-acetic acid
- TIP41:
-
PPA2 activator (tonoplast intrinsic protein)
- PM:
-
Plasma membrane
- SAL:
-
Salicilic acid
References
Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K (1997) Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9:1859–1868
Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15:63–78
Alsterfjord M, Sehnke PC, Arkell A, Larsson H, Svennelid F, Rosenquist M et al (2004) Plasma membrane H+-ATPase and 14-3-3 isoforms of Arabidopsis leaves: evidence for isoform specificity in the 14-3-3/H+-ATPase interaction. Plant Cell Physiol 45:1202–1210
Arango M, Gevaudant F, Oufattole M, Boutry M (2003) The plasma-membrane proton pump ATPase: the significance of gene subfamilies. Planta 216:355–365
Axelsen KB, Venema K, Jahn T, Baunsgaard L, Palmgren MG (1999) Molecular dissection of the C-terminal regulatory domain of the plant plasma membrane H+-ATPase AHA2: mapping of residues that when altered give rise to an activated enzyme. Biochemistry 38(22):7227–7234
Baltimore D (1985) Retroviruses and retrotransposons: the role of reverse trancriptase in shaping the eukaryotic genome. Cell 40:481–482
Baxter I, Tchieu J, Sussman MR, Boutry M, Palmgren MG, Gribskov M et al (2003) Genomic comparison of P-type ATPase ion pumps in Arabidopsis and rice. Plant Physiol 132:618–828
Baxter I, Young JC, Armstrong G, Foster N, Bogenschutz Cordova T et al (2005) A plasma membrane H+-ATPase is required for the formation of proanthocyanidins in the seed coat endothelium of Arabidopsis thaliana. PNAS 102:2649–2654
Bock KW, Honys D, Ward JM, Padmanaban S, Nawrocki EP, Hirschi KD et al (2006) Integrating membrane transport with male gametophyte development and function through transcriptomics. Plant Physiol 140:1151–1168
Buch-Pedersen MJ, Pedersen BP, Veierskov B, Nissen P, Palmgren MG (2009) Protons and how they are transported by proton pumps. Eur J Physiol 457:573–579
Chang C, Hu Y, Sun S, Zhu Y, Ma G, Xu G (2009) Proton pump OsA8 is linked to phosphorus uptake and translocation in rice. J Exp Bot 60:557–565
Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucl Acids Res 16(22):10881–10890
Davuluri RV, Sun H, Palaniswamy SK, Matthews N, Molina C, Kurtz M, Grotewold E (2003) AGRIS: Arabidopsis gene regulatory information server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinform 4:25
Duby G, Boutry M (2009) The plant plasma membrane proton pump ATPase a highly regulated P-type ATPase with multiple physiological roles. Eur J Physiol 457:645–655
Ewing N, Bennett A (1994) Assessment of the number and expression of P-Type H+ATPase genes in tomato. Plant Physiol 106:547–557
Frías I, Caldeira MT, Pérez-Castiiieira JR, Navarro-Aviiió JP, Culiaiiez-Macia FA, Kuppinger O et al (1996) A major isoform of the maize plasma membrane H+-ATPase: characterization and induction by auxin in coleoptiles. Plant Cell 8:1533–1544
Fuglsang AT, Tulinius G, Cui N, Palmgren MG (2006) Protein phosphatase 2A scaffolding subunit A interacts with plasma membrane H+-ATPase C-terminus in the same region as 14-3-3 protein. Physiol Plant 128:334–340
Fuglsang AT, Guo F, Cuin T, Qiu Q, Song C, Kristiansen K et al (2007) Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+ ATPase by preventing interaction with 14-3-3 protein. Plant Cell 19:1617–1634
Fuglsang AT, Kristensen A, Cuin TA, Schulze WX, Persson J, Thuesen KH et al (2014) Receptor kinase-mediated control of primary active proton pumping at the plasma membrane. Plant J 80(6):951–964
Gaxiola R, Palmgren MG, Schumacher K (2007) Plant proton pumps. FEBS Lett 581:2204–2214
Haberer G, Mader MT, Kosarev P, Spannagl M, Yang L, Klaus FX (2006) Mayer large-scale cis-element detection by analysis of correlated expression and sequence conservation between Arabidopsis and Brassica oleracea. Plant Physiol 142(4):1589–1602
Harper J, Manney L, DeWitt N, Sussman M (1994) The plasma membrane H+-ATPase gene family in Arabidopsis: genomic sequence of AHA10 which is expressed primarily in developing seeds. Mol Gen Genet 244:572–587
Hentzen AE, Smart LB, Wimmers LE, Fang H, Schroeder JI, Bennett AB (1996) Two plasma membrane H+-ATPase genes expressed in guard cells of Vicia faba are also expressed throughout the plant. Plant Cell Physiol 37:650–659
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res 1:297–300
Houlne G, Boutry M (1994) Identification of an Arabidopsis thaliana gene encoding a plasma membrane H+-ATPase whose expression is restricted to anther tissues. Plant J 5:311–317
Huang S, Li R, Zhang Z et al (2009) The genome of the cucumber, Cucumis sativus L. Nat Genet 41:1275–1281
Janicka-Russak M, Klobus G, Mlodzinska E, Klobus G (2009) The role of polyamines in the regulation of the plasma membrane and the tonoplast proton pumps under salt stress. J Plant Physiol 167:261–269
Janicka-Russak M, Kabala K, Wdowikowska A, Klobus G (2011) Response of plasma membrane H+-ATPase to low temperature in cucumber roots. J Plant Res 125:291–300
Janicka-Russak M, Kabala K, Wdowikowska A, Klobus G (2014) Modification of plasma membrane proton pumps in cucumber roots as an adaptation mechanism to salt stress. J Plant Physiol 170:915–922
Kabala K, Janicka-Russak M, Burzynski M, Klobus G (2008) Comparison of heavy metal effect on the proton pumps of plasma membrane and tonoplast in cucumber cells. J Plant Physiol 165:278–288
Kalampanayil BD, Wimmers LE (2001) Identification and characterization of a salt-stressed-induced plasma membrane H+-ATPase in tomato. Plant Cell Environ 24:999–1005
Kaplan B, Davydov O, Knight H, Galon Y, Knight MR, Fluhr R, Fromm H (2006) Rapid transcriptome changes induced by cytosolic Ca2+ transients reveal ABRE-related sequences as Ca2+-responsive cis elements in Arabidopsis. Plant Cell 18:2733–2748
Kasamo K (2003) Regulation of plasma membrane H+-ATPase activity by membrane environment. J Plant Res 116:517–523
Klobus G (1990) Nitrate uptake and activity of plasmalemma associated ATPase in Cucumis sativus L. roots. Acta Physiol Plant 12:225–231
Klobus G, Janicka-Russak M (2007) Modification of plasma membrane and vacuolar H+-ATPase in response to NaCl and ABA. J Plant Physiol 164:295–302
Kozak M (1991) An analysis of vertebrate mRNA sequences: intimations of translational control. J Cell Biol 115:887–903
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H et al (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Lefebvre B, Arango M, Oufattole M, Crouzet J, Purnelle B, Boutry M (2005) Identification of a Nicotiana plumbaginifolia plasma membrane H+-ATPase gene expressed in the pollen tube. Plant Mol Biol 58:775–787
Lillo C, Kataya AR, Heidari B, Creighton MT, Nemie-Feyissa D, Ginbot Z et al (2014) Protein phosphatases PP2A, PP4 and PP6: mediators and regulators in development and responses to environmental cues. Plant Cell Environ 37(12):2631–2648
Loots GG, Locksley RM, Blankespoor CM, Wang ZE, Miller W, Rubin EM et al (2000) Footprinting of the spinach nitrite reductase gene promoter reveals the preservation of nitrate regulatory elements between fungi and higher plants. Science 288:136–140
Lukaszewicz M, Jerouville B, Boutry M (1998) Signs of translational regulation within the transcript leader of a plant plasma membrane H+-ATPase gene. Plant J 14:413–423
Michelet B, Boutry M (1995) The plasma membrane H+-ATPase. A highly regulated enzyme with multiple physiological function. Plant Physiol 108:1–6
Migocka M, Papierniak A (2010) Identification of suitable reference genes for studying gene expression in cucumber plants subjected to abiotic stress and growth regulators. Mol Breed 28:343–357
Mito N, Wimmers LE, Bennett AB (1996) Sugar regulates mRNA abundance of H+-ATPase gene family members in tomato. Plant Physiol 112:1229–1236
Mlodzinska E, Wdowikowska A, Klobus G (2010) Identification and characterization of two genes encoding plasma membrane H+-ATPase in Cucumis sativus L. Acta Physiol Plant 32:1103–1111
Moriau L, Michelet B, Bogaerts P, Lambert L, Michel A, Oufattole M, Boutry M (1999) Expression analysis of two gene subfamilies encoding the plasma Membrane H+-ATPase in Nicotiana plumbaginifolia reveals the major transport functions of this enzyme. Plant J 19:31–41
Morsomme P, Boutry M (2000) The plant plasma membrane H+-ATPase: structure, function and regulation. Biomembranes 1465:1–16
Morsomme P, Kerchove d’Exaerde A, Meester SD, Thinès D, Goffeau A, Boutry M (1996) Single point mutations in various domains of a plant plasma membrane H(+)-ATPase expressed in Saccharomyces cerevisiae increase H(+)-pumping and permit yeast growth at low pH. EMBO J 15:5513–5526
Morsomme P, Dambly S, Maudoux O, Boutry M (1998) Single point mutations distributed in 10 soluble and membrane regions of the Nicotiana plumbaginifolia plasma membrane PMA2 H+-ATPase activate the enzyme and modify the structure of the C-terminal region. J Biol Chem 273(52):34837–34842
Nakajima N, Saji H, Aono M, Kondo N (1995) Isolation of cDNA for a plasma membrane H+-ATPase from guard cells of Vicia faba L. Plant Cell Physiol 36:919–924
Nicolic M, Cesco S, Monte R, Tomasi N, Gottardi S, Zamboni A, Pinton R, Varanini Z (2012) Nitrate transport in cucumber leaves is an inducible process involving an increase in plasma membrane H+-ATPase activity and abundance. BMC Plant Biol 12:66
Niu X, Narasimhan ML, Salzman RA (1993) NaCl regulation of plasma membrane H+-ATPase gene expression in Glycophyte and Halophyte. Plant Physiol 103:712–718
Ouffatole M, Arango M, Boutry M (2000) Identification and expression of tree new Nicotiana plumbaginifolia genes which encode isoforms of a plasma-membrane H+-ATPase, and one of which is induced by mechanical stress. Planta 210:715–722
Palmgren MG (2001) Plant plasma membrane H+-ATPases powerhouses for nutrient uptake. Annu Rev Plant Physiol Plant Mol Biol 52:817–845
Pasquier C, Promponas VJ, Palaios GA, Hamodrakas JS (1999) A novel method for predicting transmembrane segments in proteins based on a statistical analysis of the SwissProt database: the PRED-TMR algorithm. Protein Eng 12:381–385
Perez C, Michelet B, Ferrant V, Bogaerts P, Boutry M (1992) Differential expression within a three-gene subfamily encoding a plasma membrane H+-ATPase in Nicotiana Plumbaginifolia. J Biol Chem 267:1204–1211
Pii Y, Alessandrini M, Guardini K, Zamboni A, Varanini Z (2014) Induction of high-affinity NO3− uptake in grapevine roots is an active process correlated to the expression of specific members of the NRT2 and plasma membrane H+-ATPasegene families. Funct Plant Biol 41:353–365
Portillo F (2000) Regulation of plasma membrane H+-ATPase in fungi and plants. BBA 1469:31–42
Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R (2005) InterProScan: protein domains identifier. Nucleic Acids Res 33(Web Server issue):W116–W120. doi:10.1093/nar/gki442
Rastogi R, Bate NJ, Sivasankar S, Rothstein SJ (1997) Footprinting of the spinach nitrite reductase gene promoter reveals the preservation of nitrate regulatory elements between fungi and higher plants. Plant Mol Biol 34:465–476
Reineke AR, Bornberg-Bauer E, Gu J (2011) Evolutionary divergence and limits of conserved non-coding sequence detection in plant genomes. Nucleic Acids Res 39:6029–6043
Robertson WR, Clark K, Young JC, Sussman MR (2004) An Arabidopsis thaliana plasma membrane proton pump is an essential for pollen development. Genetics 168:1677–1687
Santi S, Locci G, Monte R, Pinton R, Varanini Z (2003) Induction of nitrate uptake in maize roots: expression of a putative high-affinity nitrate transporter and plasma membrane H+-ATPase isoforms. J Exp Bot 54:1851–1864
Santi S, Cesco S, Varanini Z, Pinton R (2005) Two plasma membrane H+ATPase genes are differentially expressed in iron-deficient cucumber plants. Plant Physiol Biochem 43:287–292
Sondergaard TE, Schulz A, Palmgren MG (2004) Energization of transport processes in plants. Roles of the plasma membrane H+-ATPase. Plant Physiol 136:2475–2482
Stewart AJ, Hannenhalli S, Plotkin JB (2012) Why transcription factor binding sites are ten nucleotides long. Genetics 192:973–985
Tamura K, Dudley J, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 10:1093
Tusnady GE, Simon I (2001) The HMMTOP transmembrane topology prediction server. Bioinformatics 17:849–850
Virart V, Baxter Doerner IP, Harper JF (2001) Evidence for a role in growth and salt resistance of plasma membrane H+ATPase in root endodermis. Plant J 27:191–201
Woycicki R, Witkowicz J, Gawronski P, Dabrowska J, Lomsadze A (2011) The genome sequence of the North-European cucumber (Cucumis sativus L.) unravels evolutionary adaptation mechanisms in plants. PLoS One 6(7):e22728
Yan F, Zhu Y, Muller C, Zorb C, Schubert S (2002) Adaptation of H+-pumping and PM H+-ATPase activity in proteoid roots of white lupin under phosphate deficiency. Plant Physiol 129:50–63
Yanagisawa S (2004) Dof domain proteins: plant-specific transcription factors associated with diverse phenomena unique to plants. Plant Cell Physiol 45:386–391
Zhao R, Dielen V, Kinet JM, Boutry M (2000) Cosuppression of plasma membrane H+-ATPase isoform impairs sucrose translocation, stomatal opening, plant growth, and male fertility. Plant Cell 12:535–546
Zhu Y, Di T, Xu G, Chen X, Zeng H, Yan F, Shen Q (2009) Adaptation of plasma membrane H(+)-ATPase of rice roots to low pH as related to ammonium nutrition. Plant Cell Environ 32:1428–1440
Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136(1):2621–2632. http://www.genevestigator.org
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by E. Schleiff.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11738_2016_2152_MOESM1_ESM.tif
Supplementary material 1 (TIFF 544 kb) Figure S1. MultiAlin alignment of CsHA1 - CsHA10. High consensus amino acids are indicated with red while low consensus residues in blue; black- neutral consensus color. The black boxes mark the conserved sequences common to P-ATPases: (TGES, DKTGTLT, GDGVNDA and KGAP). The putative transmembrane domains (TMDs) are sign with green boxes
11738_2016_2152_MOESM2_ESM.tif
Supplementary material 2 (TIFF 6221 kb) Figure S2. Relative expression of CsHA2, sHA3, CsHA4, CsHA8, CsHA9 and CsHA10 in roots and leaves of 14-day-old cucumbers growing under control conditions. Expression levels measured with qPCR are given as percentage of internal standard (TIP41) copies. Error bars represent SD. For each of the two independent RNA extractions, measurements of gene expression were obtained in duplicate or triplicate
11738_2016_2152_MOESM3_ESM.tif
Supplementary material 3 (TIFF 1582 kb) Figure S3. Expression of CsHA1 measured with RT-PCR in cucumber roots and leaves of 2-week-old cucumber under hormones and stress factors. The 14-day-old plants growing in standard nutrient solution were transferred to 50 μM ABA, IAA or GA3, 100 μM salicylic acid or 150 μM ACC solutions for 5 h. The effect of NaCl, PEG and light conditions was tested after 24 h exposure of 13-day-old plants to stress factors, whereas in experiments on different nutrient supplies, 9-day-old cucumber seedlings were transferred for 5 days to the nutrient solution without -N, -P or -S. The experiment was repeated three times independently with comparable results
Rights and permissions
About this article
Cite this article
Wdowikowska, A., Klobus, G. The plasma membrane proton pump gene family in cucumber. Acta Physiol Plant 38, 135 (2016). https://doi.org/10.1007/s11738-016-2152-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11738-016-2152-4