Abstract
MicroRNAs regulate gene expression by guiding the cleavage or attenuating the translation of target mRNAs. In Arabidopsis thaliana, the subset of class II TCP genes (plant-specific group of transcription factors) contains an miR319-binding site. One of them, AtTCP4, regulates negatively leaf growth and positively leaf senescence. In addition, miR319 targeting of TCP4 is critical for petal and stamen development and affects flowering time. The aim of this work was to identify the cDNA of InTCP4 gene and In-miR319 precursor in Ipomoea nil (Pharbitis nil). The cDNA sequence of InTCP4 shows a significant similarity to the cDNA members of the TCP family of other plant species and contains nucleotides complementary to miR319. The identified sequence In-pre-miR319 creates a long hairpin structure and mature miRNA sequence is located in a similar place as in precursors found in other plant species. Accumulation of InTCP4 mRNA and In-pre-miR319 was examined in various organs of I. nil plants. We found that the InTCP4 is strongly expressed in cotyledons of I. nil seedlings while the In-pre-miR319 accumulates mainly in the hypocotyls of seedlings. Moreover, we investigate the role of InTCP4 in the flowering induction, flower development and cotyledon senescence in I. nil. We indicate that the InTCP4 expression is controlled by both light/clock and miR319. Both InTCP4 and InMIR319 probably participate in the regulation of such processes as do their homologues in other plant species, the development of cotyledons, leaves and flower elements. The main function of InMIR319 seems to be the regulation of InTCP4 organ localization.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
MicroRNAs about 19–21 nt in length are involved in silencing target genes, by inactivation of their expression in plants usually at the transcriptional level. MiRNAs are obtained as a result of MIR gene expression encoding precursor RNA, which is sliced to the mature miRNA molecule. The miRNA nucleotide sequence length of 19–21 nt is sufficiently long to bind complementary-specific mRNA encoding some protein, and lead to its degradation or inhibit the process of translation. What is more, the miRNA sequence and the complementary sequence in the target mRNA are highly conserved within kingdoms (Reinhart et al. 2002; Zhang et al. 2007; Voinnet 2009).
On the basis of research conducted on Arabidopsis thaliana, a number of miRNAs involved in the regulation of various developmental processes were identified (Bonnet et al. 2006; Jones-Rhoades et al. 2006; Rubio-Somoza and Weigel 2011). Among others, it has been shown that the miRNA and its target genes regulate different stages of development of leaves, from the initiation of their formation to senescence (Kidner 2010; Pulido and Laufs 2010).
Leaf development is a comprehensive process that involves the transition from a small group of undifferentiated cells to a structure of a particular size and shape, highly organized, composed of different cell types with specialized functions. During this developmental sequence of events, growth and differentiation must be coordinated by a complex regulatory network in which small RNAs, mainly microRNA, but also ta-siRNAs (trans-acting small interfering RNA), begin to play an important role as a new class of gene expression regulators (Kidner 2010; Pulido and Laufs 2010). The main function of leaves is to carry out the process of photosynthesis and transpiration; they are the sites of intensive gas exchange between the plant and the environment as well. The leaves and cotyledons are also places of photoperiodic signal perception in the photoperiodic flowering induction, where the formed flowering signal is transported to the apical meristem, which leads to the transformation of vegetative shoot apical meristem into generative (Thomas and Vince-Prue 1997; Izawa et al. 2002; Hayama and Coupland 2004; Imaizumi and Kay 2006).
In the mechanism of formation of the leaves, the miR160 target genes are ARF10 (AUXIN RESPONSE FACTOR), ARF16 and ARF17 (Mallory et al. 2005; Wang et al. 2005; Liu et al. 2007). Next, miR164 regulates the size of the border between the organs by modulating expression of CUC1 (CUP-SHAPED COTYLEDONI1) and CUC2 (Laufs et al. 2004; Mallory et al. 2004; Baker et al. 2005; Sieber et al. 2007). Two miRNAs, miR156 and miR172, are involved in the phase transition from juvenile to mature plants of A. thaliana and Zea mays (Aukerman and Sakai 2003; Lauter et al. 2005; Wu et al. 2009). In the process of leaf senescence in A. thaliana two miRNAs were described: miR164 and miR319, for which the target genes are ORE1 and TCP2, -3, -4, -10, -24, respectively (Woo et al. 2001; Schommer et al. 2012). All targets of miR319 are closely related members of the class II subclass of TCP genes (Palatnik et al. 2003). In plants, the genes encoding TCP-specific transcription factors containing a conserved DNA-binding domain named TCP. This domain does not show, at the level of sequence, any homology to other known DNA-binding domains, but forms a structure similar to the bHLH domain (basic Helix–Loop–Helix). Such a domain protein is described in TB1 (TEOSINTE BRANCHED1) from maize, CYC (CYCLOIDEA) of Antirrhinum and PCF1 and 2 (PCNA PROMOTER BINDING FACTOR) of rice, hence the name of this domain (Cubas et al. 1999; Martin-Trillo and Cubas 2010).
Ipomoea nil (called also Pharbitis nil) is one of the most commonly used model short day plants (SDP) to study the process of photoperiodic flower induction. Treatment of I. nil seedlings at an early stage of development with a single cycle of induction (16 h of darkness) is sufficient for the plants to produce generative buds (Vince-Prue and Gressel 1985; Hayama et al. 2007). The place of perception of the photoperiodic signal may be in cotyledons of seedlings as young as 5 days old. This is the time when these organs have the highest sensitivity to the photoperiodic night. This is probably related to the achievement by the cotyledons of the appropriate stage of development (Ogawa and King 1990; Hayama et al. 2007).
In our previous paper, we described the first miR172 target gene InAP2-like identified in I. nil which is involved in the regulation of photoperiodic flower induction in I. nil (Glazińska et al. 2009). Studies indicate that in A. thaliana and tomato, miR319 and its target genes from the TCP family are involved in the regulation of both morphogenesis and leaf senescence. This probably occurs through control of the expression of JA biosynthesis genes, such as LOX2 (LIPOXYGENASE 2) (Schommer et al. 2008). More than that, the rTCP mutants of A. thaliana with an altered miRNA-binding site, and thus “resistant” to miR319 show, in addition to changes in leaf morphology, also delayed flowering. miR319a targeting of TCP4 is also critical for petal and stamen development in Arabidopsis (Schommer et al. 2012).
Numerous studies have shown that JA is involved in the regulation of many processes such as flower development, senescence and organ abscission (Ueda and Kato 1980; Turner et al. 2002; Yoshida 2003; Acosta and Farmer 2010). JA is an inhibitor of photosynthetic enzymes, inhibits the biosynthesis and accelerates the degradation of carboxylase ribulose-1,5-bisphosphate, and the rate of breathing and the proteolytic activity (Parthier 1990). JA can thus affect the “biological” age of the cotyledons, and thus adjust the time sensitivity of plants to the inductive photoperiod. The I. nil study demonstrated that disruption of the endogenous level of JA during night inhibits the induction of flowering (Maciejewska et al. 2004; Kęsy et al. 2012); therefore we identify InTCP4 cDNA and In-pre-miR319 from I. nil cotyledons, observe its expression in various organs of plants and investigate the role of InTCP4 in flowering induction. We also examine their participation in flower development and cotyledon senescence of I. nil.
Materials and methods
Plant material
Seeds of Ipomoea nil Chois cv. Violet (Marutane Seed Co., Kyoto, Japan) were treated according to Glazińska et al. (2009).
Organ expression
Plants were grown under LD conditions for 5 days and then were exposed to a 16-h-long inductive night. The apexes, cotyledons, hypocotyls and roots were picked after a short night in 6 days of growth and immediately frozen in liquid nitrogen and stored at −80 °C. In addition, pistils, stamens, sepals and petals were harvested from fully open flowers. Leaves were harvested from 3-week-old plants (Fig. 4).
Each experiment was repeated at least three times. All data are presented as mean ± standard error (SE).
Expression analysis during pistil and stamen development
For development analysis of pistils and stamens, flower buds were collected at various stages of growth (Fig. 4. Stage 1–5), from the appearance of flower buds with visible stamens and pistils to fully open flowers. Flower elements, pistils and stamens, were collected from all stages of growth. Thereafter, material was frozen in liquid nitrogen and stored at −80 °C for RNA isolation.
Expression analysis under photoperiodic conditions
The expression analysis of InTCP4 was performed in four photoperiodic variants. In all variants, the seedlings of I. nil were grown under LD conditions for 5 days and subsequently divided into two portions. The first one was left under LD conditions (8 h of darkness—negative flowering control) (variant I), the second was transferred to SD conditions (16-h night) (variant II–IV). In variant II, the seedlings were irradiated in hour 8 of the darkness with a 15-min-long pulse of red light (R, 1.5 μmol m−2 s−1, fluorescent tubes TLD 15 Red/18 W, Philips, Holland). In variant III, IAA applications on cotyledons were performed once before starting the 16-h inductive night. IAA (0.5 mM) applications to both sides of the cotyledons were performed with the use of a small soft brush (about 50 μL/plant). Variant IV was control plants without any treatment (positive flowering control).
Cotyledons (without petioles) were harvested every 3 h after transfer to the darkness (variant I and IV). In experiments with the influence of IAA and a 15-min-long pulse of red light (variant II, III) for InTCP4 expression analysis, cotyledons were picked every 3 h after hormone or light application. Subsequently, the cotyledons were frozen in liquid nitrogen and stored at −80 °C. All manipulations during the dark period were done under dim green safe light.
Expression analysis during cotyledon senescence
The 6-day-old plants were divided into four variants. Three subjected to ageing, which was induced by placing a dark (variant I) or exogenous application of phytohormones, accelerating the ageing process, such as jasmonic acid methyl ester (JAMe) at a concentration of 100 μM (variant II), and abscisic acid (ABA) with a concentration of 0.5 mM (variant III). Some plants were left at LD for further growth as a control (variant IV). Samples were collected in the next 6 days of growth after the phytohormone application or inserted into the darkness. Before starting the experiment, and at each harvest, to monitor the ongoing process of ageing, the amount of chlorophyll in the cotyledons was investigated. The material collected was placed immediately in liquid nitrogen and stored at −80 °C.
Molecular cloning of InTCP4 cDNA
The tissue of I. nil (1.0–1.5 g) was homogenized in a sterile chilled mortar with a pestle. Total RNA was isolated from cotyledons with the GeneMATRIX Universal RNA Purification Kit (EurX Gdansk, Poland) according to the manufacturer’s instructions. First-strand cDNA was synthesized with 1 μg of total RNA and 0.5 μg anchored oligo (dT)19 primer according to the protocol Transcriptor High Fidelity cDNA Synthesis Kit (ROCHE Diagnostics GmbH, Germany). PCR, using degenerated primers 5′-WGTYGARGTRCAAGGYGGTCACAT-3′ (forward) and 5′- AGGWGTGTWACTGGACTGAAGGGGW-3′ (reverse), was performed in the T3 Thermocycler (Biometra, Göttingen, Germany). Primers were designed from sequences conserved between TCP4 from A. thaliana, Solanum lycopersicum and Malus domestica. All primers used were synthesized by Genomed S.A. Warsaw, Poland. Mixtures and conditions of PCR reaction were prepared according to Glazińska et al. (2009). An amplified cDNA fragment was isolated from an agarose gel with the GeneMATRIX Agarose Out DNA Purification Kit (EurX, Gdansk, Poland), cloned using the TOPO TA Cloning Kit (Invitrogen, Carlsbad, California) and sequenced by Genomed S.A. Warsaw, Poland. A full-length cDNA coding for InTCP4 was isolated using the BD SMART RACE cDNA Amplification Kit (Clontech-Takara Bio Europe, Saint-Germain-en-Laye, France). mRNA used in 5′- and 3′-RACE-PCR was isolated with Dynabeads Oligo (dT)25 (Dynal Biotech, Oslo, Sweden). Gene-specific primers for 5′-RACE (5′-AGCAGCTTCAGCAACAGGAGGATACTTG-3′ outer primer, 5′-GGCACGGTGGTGGTGGTGGAGACG-3′ inner primer) and 3′-RACE (5′-CGTAGCCGAGCCGATCCTGGACATC-3′ outer primer, 5′-GGGTTAACGGTACTGGGTTTCCAGGCG-3′ inner primer) were picked from a cDNA sequence of the identified clone using FastPCR 5.4.30 software. PCR reactions were performed using the Advantage 2 PCR Enzyme System (Clontech-Takara Bio Europe, Saint-Germain-en-Laye, France). The RACE products were cloned using the TOPO TA Cloning Kit (Invitrogen, Carlsbad, California) and sequenced by Genomed S.A. Warsaw, Poland.
The identified cDNAs of the InTCP4 gene were analysed using ClustalW (http://www.ebi.ac.uk/clustalw), BLAST 2.2.25 (http://www.ncbi.nlm.nih.gov/BLAST) ExPASY (http://www.expasy.org) and Search miRBase::Sequences (http://microrna.sanger.ac.uk/sequences/). The phylogenetic analysis was made using the Phylogeny.fr web service (http://www.phylogeny.fr/version2_cgi/simple_phylogeny.cgi). Sequence alignments were carried out using the MUSCLE software and then the Gblocks program was used to eliminate poorly aligned positions and divergent regions. The phylogenetic analysis was made using the PhyML program. The phylogenetic tree was constructed using the TreeDyn program.
Identification of In-pre-miRNA319 sequence
Basic Local Alignment Search Tool (BLAST) (Altschul et al. 1990) was used to search for the fragment pri-miR319 including the pre-miRNA sequence in the EST sequences database of I. nil. The MiR319 sequence was obtained from the miRBase database (ftp://mirbase.org/pub/mirbase/12.0/) (Griffiths-Jones et al. 2008). Because sequences of miR319 show conservation across different kingdoms (Axtell and Bartel 2005; Zhang et al. 2006), we use the miRNAs from A. thaliana. EST no: CJ744218 that potentially contain miRNA sequences were obtained after the BLAST search. The RNAstructure software (http://rna.urmc.rochester.edu/RNAstructureWeb/Servers/Predict1/Predict1.html) was used to compute their secondary structure.
The presence of the identified sequence in the plant was confirmed by the RT-PCR reaction with specific primers 5′-ACTGAGTGAATGATGCGGGAGACAA-3′ (forward) and 5′-CCAATGAAAAACCCTTTCTCCAGC-3′ (reverse) with GoTaq Hot Start Colorless Master Mix (Promega Corporation, USA) according to the manufacturer’s instructions. An amplified DNA fragment was isolated from an agarose gel with the GeneMATRIX Agarose Out DNA Purification Kit (EurX, Gdansk, Poland), cloned using the TOPO TA Cloning Kit (Invitrogen, Carlsbad, CA, USA) and sequenced by Genomed S.A. Warsaw, Poland. The identified sequences were analysed using Search miRBase web software.
Quantitative RT-PCR analysis of InTCP4 gene and In-pre-miR319 expression
The gene expression analysis was performed by real-time PCR (qPCR) with a LightCycler 2.0 Carousel-Based System (ROCHE Diagnostics GmbH, Germany) and LightCycler TaqMan Master Kit (ROCHE Diagnostics GmbH, Germany). The templates for qRT-PCR were prepared the same way as for molecular cloning of InTCP4 cDNA.
Gene-specific primers and a UPL probe were designed using Universal ProbeLibrary Assay Design Center (http://www.roche-applied-science.com/sis/rtpcr/upl). The qPCR reactions were made for the following genes: InTCP4, InCAB2 (CHLOROPHYLL A/B BINDING PROTEIN2) (GenBank: D78207), InLOX2 (identified EST sequences in the NCBI database of I. nil: CJ761975), and InACT4 (GenBank: HM802138.2). Actin was used as reference endogenous control for normalization purposes. This technique was also used to analyse the accumulation of In-pre-miR319.
The qPCR conditions included an initial denaturation step 95 °C for 10 min followed by 45 cycles of 95 °C for 10 s, 58 °C for 30 s, 72 °C for 1 s followed by 1 cycle of cooling at 40 °C for 30 s.
qPCR analyses were performed with the gene-specific primers: InTCP4 (61 bp) 5′-GTTAACCCGAATTTCGACCA-3′ (forward) and 5′-CATCTCCGGGGGTCTCTT-3′ (reverse) with hydrolysis probe UPL 105, In-pre-miR319 (76 bp) 5′-TTCCCTTTTCCACACTAATCG-3′ (forward) and 5′-AACTGGCAAACACCAACAAA-3′ (reverse) with hydrolysis probe UPL20, InLOX2 (70 bp) 5′-CACGCGGACAAGAAGGAT-3′ forward) and 5′-TCAGGATTCCAATCAGGTCA-3′ (reverse) with hydrolysis probe UPL 64, InCAB2 (78 bp) 5′-CGGAAAGGCTGTGAAGGTC-3′ (forward) and 5′-CCAGCTGTCTTCCTCATGGTA-3′ (reverse) with hydrolysis probe UPL 97, InACT4 (65 bp) with the gene-specific primers 5′-GGAAATACAGTGTCTGGATTGGA-3′ (forward) and 5′-CCACATCTGTTGGAATGTGC-3′ (reverse) with hydrolysis probe UPL 139 (ROCHE Diagnostics GmbH, Germany). qPCR mixtures were made as follows: 1 μL of 1st strand cDNA, 0.4 μL of gene-specific primer solution (10 μM), 0.2 μL of gene-specific hydrolysis probe and 4 μL of Master Mix (containing reaction buffer, dNTP mix and DNA Polymerase) from LightCycler TaqMan Master (ROCHE Diagnostics GmbH, Germany). The computer application used for the analysis was LCS4.0 (ROCHE Diagnostics GmbH, Germany) and for the calculations and graphs, MS Office Excel (Microsoft). qPCR reactions were performed in triplicate for each RNA sample. All data are presented as mean ± standard error (SE).
Measurement of the relative amounts of chlorophyll in cotyledons
The amount of chlorophyll was determined by the method of CCl (chlorophyll content index) with the device Opti-Sciences CCM-200 (Opti-Sciences, Inc., USA). Each measurement of relative chlorophyll content was performed on at least three different plants of the experimental variant. Each measurement was performed three times. The data obtained were then subjected to statistical analysis.
Mapping of the InTCP4 mRNA internal cleavage sites
To determine the internal cleavage site in InTCP4 mRNA, RNA ligase-mediated rapid amplification of cDNA ends (5′ RACE) was performed with the First Choice RLM-RACE Kit (Ambion). The procedure was based on Glazińska et al. (2009). A unique gene-specific DNA fragment was first amplified with the 5′ RACE outer primer (Ambion) and gene-specific primer (5′-TACCTCGGGTGACCGCCATTGG-3′) and then with the 5′ RACE inner primer (Ambion) and nested gene-specific primer (5′-AGCAGAGGATGGCTTGTCGGA-3′). The RACE products were cloned using the TOPO TA Cloning Kit (Invitrogen, Carlsbad, CA, USA) and sequenced by Genomed S.A. Warsaw, Poland.
Results
Isolation of cDNA for the InTCP4 gene
The cDNA sequence of the gene TCP4 homologue was identified with RT-PCR with degenerate primers. These primers were designed to conserve sequences encoding the TCP domain, and a sequence complementary to miR319 based on cDNA sequence comparisons of the TCP gene family present in different plant species. After the RT-PCR reaction, a single product, with a molecular weight of 800 bp, was obtained (data not shown). The obtained reaction products were isolated, cloned and sequenced. Comparison of the sequenced cDNA fragment with the sequences chosen by BlastN showed its highest homology to cDNAs of the TCP gene family from other plant species. To isolate the full cDNA sequence of the gene InTCP4, the 5′ and 3′ RACE method was used. The obtained reaction products were isolated and cloned in the same way as for the reaction with degenerate primers. After connection all identified fragments formed a full-length cDNA gene InTCP4, 1,523 bp in length (Fig. 1a). Then, the sequence was deposited in the NCBI database (National Center for Biotechnology Information) as I. nil TCP family transcription factor TCP4 mRNA, complete cds no: HM357790.
a Nucleotide and deduced amino acid sequence of InTCP4. Amino acid sequences marked in grey represent TCP domain. Underlined nucleotide sequences correspond to target site for miR319. b The phylogenetic relationship of InTCP4 compared with the TCP genes family from various plant species. GeneBank Accession Nos. from top to bottom HM210875.1, AF175965.1, HM210876.1, EF091574.1, AY205603.1, AB531021.1, NM_180258.2, GQ505958.1. c Alignment of part of nucleotide sequences of InTCP4 with homologous genes from other plant species and mature miR319a, -b, and -c from A. thaliana. Stars denote identical nucleotides
The deduced InTCP4 protein had 400 aa residues and possesses TCP domains, characteristic for transcriptional factors from the TCP family (Fig. 1a).
An analysis of phylogenetic relationships between the identified cDNA of the InTCP4 sequence and the cDNAs of the TCP gene family revealed that isolated cDNA was very closely related to CYCLOIDA (CYC) from Lycopersicon esculentum, and LANCEOLATE (LA) from Capsinum annum and Solanum melongena. A high degree of similarity of the sequence was also found for cDNA CINCINNATA (CIN) Antirrhinum majus, TCP4A Malus domestica, and TCP4 A. thaliana (Fig. 1b). Analysis of InTCP4 cDNA using the software available on http://microrna.sanger.ac.uk/sequences/ also revealed the sequence complementary to miR319. This sequence is located at the end of the protein coding sequence, as in the other TCP genes identified in other plant species (Fig. 1c).
Identification of In-pre-miRNA319 sequence
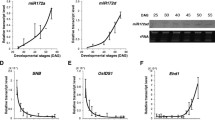
To identify the precursor of miR319, the I. nil EST sequence database was searched with the miR319 sequence derived from A. thaliana. The BlastN program indicated that SEQ ID CJ744218 showed high homology to the mature and precursor molecule of miR319 of other plant species (data not shown). We confirmed the presence of the identified sequence in plants using the RT-PCR reaction. An amplified DNA fragment was sequenced and analysed for the presence of sequences of mature miR319 (Fig. 2a) and secondary structure (Fig. 2b) with Search miRBase::Sequences and RNAstructure software, respectively. A putative sequence of the mature molecule 21 nt miR319a/b occurs between 389 and 409 bp and is located in the stem forming a hairpin structure (Fig. 2a, b). The identified sequence was compared with the stem-loop pre-miR319 sequences of other plant species and shows great similarity to the miR319 precursor sequence of a number of plant species, the highest to Cucumis melo, Hevea brasiliensis and Ricinus communis (Fig. 2c).
Alignment of the nucleotide sequences of In-pre-miR319 with a mature miR319a, -b, and -c from A. thaliana, b precursors of miR319 of other plant species indicate the highest homology. Stars denote identical nucleotides. miRBase Accession Nos. from top to bottom MI0013406, MI0023233, MI0022056. c Secondary stem-loop structures of the In-pre-miR319. Localization of mature miR319 is indicated on the stem of the precursor sequence
MiR319 mediates cleavage of InTCP4 mRNA
The specific cleavage site of the InTCP4 mRNA was determined by 5′ RACE, which takes advantage of the monophosphate present at the 5′ terminus of the 3′ cleavage fragment (Kasschau et al. 2003). The distinct PCR band of the expected size was observed for the InTCP4 target (Fig. 3). Cloning and DNA sequencing of this amplified product mapped the 5′ end of the cleavage products to the designated position at the middle of the region complementary to miR319.
5′- RACE to determine the cleavage site. a Nested PCR product (line InTCP4), spanning between the primer and cleavage sites, was used for cloning into a vector for DNA sequencing. Lane M represents 100 bp DNA ladders. b The InTCP4 mRNA cleavage site is designated at the 11th base from the 5′ end, indicated by the arrow
Expression patterns of the InTCP4 and MIR319 genes in various organs of I. nil
Accumulation of InTCP4 and MIR319 mRNA was examined in the vegetative and generative tissues of seedlings and mature I. nil plants (Fig. 4). The highest levels of InTCP4 transcripts were indicated in cotyledons and next in shoot apices of 6-day-old seedlings. The high level of expression was also observed in pistils. A much lower level of mRNA InTCP4 was observed in petals, sepals and leaves (Fig. 5a).
Development of I. nil seedlings and flowers. a 5- day-old seedling. b Shoot apices of 5-day-old seedling. c Leaf of mature plant. d Development of flowers. Upper panel—whole flower buds, below—buds with flower elements shown. On the right—flower elements of a fully open flower. Numbers (1–5) represent the stages of flower growth from the first flower bud with easily visible stamens and pistils to fully open flowers. The symbols are: C cotyledons, H hypocotyls, R roots, SA shoot apices, L leaf, ST stamen, PI pistil, P petal, S sepals. Black line—bar 1 cm
The level (related to InACT4) in various plant tissues of I. nil, a InTCP4 transcript and b In-pre-miR319 accumulation. The symbols are: C cotyledons, H hypocotyls, R roots, SA shoot apices, L leaf, ST stamen, PI pistil, P petal, S sepals. c The transcript level of InTCP4 (related to InACT4) during stamen and pistil development of I. nil. Numbers (1–5) represent the stages of flower growth from the first flower bud with easily visible stamens and pistils to fully open flowers
The results of RT-qPCR showed that In-pre-miR319 accumulates mainly in the hypocotyls of seedlings. In other organs, except shoot apexes of seedlings, the level of In-pre-miR319 was very low or almost absent (Fig. 5b).
Analysis of transcript level of InTCP4 during pistil and stamen development
For analysis of pistil and stamen development, flower elements were isolated from generative buds collected at various stages of growth (Fig. 4), from the appearance of flower buds with easily visible stamens and pistils (stages 1–4) to fully open flowers (stage 5).
The transcript level of InTCP4 increases during pistil growth (Fig. 5c). During the growth of stamens, InTCP4 expression pattern is opposite and the lowest level occurs in the stamens of open flowers (Fig. 5c).
InTCP4 expression under LD and SD conditions
To investigate the possible participation of InTCP4 in the mechanism of flower induction in I. nil, gene expression was examined in cotyledons harvested during LD (8-h darkness/16-h light) and SD conditions (16-h darkness/8-h light) (Fig. 6).
The level (related to InACT4) of InTCP4 transcript (three replicates) in the cotyledons of plants cultivated under different light conditions and treated with IAA. The symbols are: LD long day; SD short day; SD + IAA—SD and IAA treatment before transfer to darkness; SD + NB—16-h darkness broken (night break) with 15 min red light. Black lines—dark period; arrows indicate IAA application to cotyledons and 15 min of red light
We observed that the level of InTCP4 transcript successively decreased during the night. In light, a slight temporary increase in transcriptional activity occurred followed by a decrease and then gradual increase in expression during the day. For LD conditions, the amount of mRNA went back to the level before the night (Fig. 6). We also investigated the effect of factors that repress the flowering induction of I. nil on expression of InTCP4. As inhibitory factors, we used the IAA treatment before transfer of seedlings to inductive darkness and a night break in the 8th h with 15 min of red light in the 16-h inductive night. The obtained results indicated that auxin does not affect the expression of the gene. A night break causes a short-term increase in InTCP4 expression and then a decrease in a similar way as in SD conditions (Fig. 6).
The investigation of the possible role of InTCP4 in cotyledon senescence of I. nil
To investigate the possible role of InTCP4 in cotyledon senescence of I. nil, we analysed the expression of InTCP4 and jasmonic acid biosynthesis gene (InLOX2) during the various stages of ageing (Fig. 7c, d). As ageing-inducing factors, continuous darkness, jasmonic acid methyl ester (JAMe) and abscisic acid (ABA) were used. Changes in the amount of chlorophylls and InCAB2 gene expression were used in the experiment as indicators of the ongoing process of senescence (Fig. 7a, b).
All the factors that induce ageing caused a decrease in the relative content of chlorophyll, and the expression of InCAB2. The strongest decrease was observed under the influence of darkness. In plants grown under control conditions (LD), a slight decrease was observed on the tested parameters due to the presence of the natural ageing process of the cotyledons, as a result of the appearance of the leaves on the 8th day of growth of seedlings. InLOX2 gene transcription activity in plants grown in the light and in the dark remained at a low level. The application of ABA and JAMe promotes the expression of InLOX2; however, the stimulatory effect of phytohormones is getting weaker in the days of the experiment (Fig. 7c). In cotyledons of the plants treated with ABA and JAMe compared to those grown in the light (control) we observed a slight increase in the level of transcript InTCP4, while darkness causes a significant decrease in the expression of a gene (Fig. 7d).
Discussion
Representatives of the class of TCP genes are important regulators of plant growth and development. They participate in the creation of the proper symmetry of flowers and petals, plant architecture, leaf morphogenesis, senescence, and regulate processes such as embryonic growth and circadian rhythm (Navaud et al. 2007; Giraud et al. 2010; Martin-Trillo and Cubas 2010). The activity of the part of the TCP gene family in A. thaliana is controlled by the family of MIR genes encoding regulatory micro RNA molecules—miR319 (Schommer et al. 2012).
To identify the InTCP4 cDNA in I. nil plants, the RT-PCR technique and degenerated primers were used. Primers were designed for the sequence encoding DBD domain, as well as for the sequence containing nucleotides complementary to miR319. Using a 3′ and 5′ RACE-PCR reaction, we obtained the full sequence of InTCP4 cDNA. The complete cDNA (GenBank accession number EF216864) consists of 1,523 bp and encodes 400 amino acids (Fig. 1a). An analysis of phylogenetic relationships between the identified cDNA of the InTCP4 sequence and the cDNAs of the TCP gene family revealed that isolated cDNA was very closely related to CYC from L. esculentum, LA from C. annum and S. melongena. A relatively high degree of homology was also indicated for CIN A. majus, TCP4A M. domestica, and TCP4 A. thaliana (Fig. 1b). Comparison between InTCP4 mRNA and miR319a, -b, -c, -d from A. thaliana indicates that the isolated cDNA sequence contains miR319-complementary sequence and the strongest complementary sequence occurs to miR319c (Fig. 1c). The miRNA target site is located outside the TCP domain, near the 3′ part of the coding region, as in the other TCP genes identified in other plant species (Fig. 1c). The interaction between the InTCP4 and miR319, as in other plant species (Schommer et al. 2012), has up to six mismatches, which is higher than in other known plant miRNA target pairs (Fig. 1c) but still, the predicted free energy is good enough for efficient interaction and guiding cleavage of the target mRNA (Palatnik et al. 2003). A direct role for miR319 in reducing InTCP4 mRNA levels was demonstrated by the identification of InTCP4 cDNA fragments whose 5′ termini are consistent with the products expected from miR319-directed mRNA cleavage (Fig. 3).
Members of the TCP family share an approximately 60-residue-long TCP domain that binds to DNA. Secondary structure prediction revealed that this domain forms a basic region followed by two helices separated by a loop (Cubas et al. 1999; Kosugi and Ohashi 2002; Aggarwal et al. 2010), even though it shares little sequence similarity with the classical bHLH domain.TCP domain class II starts from an N-terminal aminoacid Lys (K), which is conserved in 93 % of the members of the TCP protein family and contains three–residue insertion in the basic domain that is absent from class I (Aggarwal et al. 2010). Analysis of the putative amino acid InTCP4 sequence showed the presence of Lys and all other amino acids required for creating a functional TCP domain class II (Fig. 1a). This TCP domain is most similar to the domain present in CIN A. majus (data not shown), which indicates that InTCP4 may take part in the regulation of similar processes. The CIN gene is responsible for regulating the growth of the leaf blade and influencing the morphogenesis of flower petals (Crawford et al. 2004). Similar functions are also played by another gene with high similarity to the nucleotide sequence InTCP4 of I. nil, namely LA from S. melongena. The LA gene regulates development of the leaves, and is also a target gene for MIR319 (Ori et al. 2007).
To compare the expression of InTCP4 and MIR319, we decided to identify the sequence of miR319 precursor. Because the sequence of miR319 showed conservation across different plant species (Zhang et al. 2006) we use the A. thaliana miR319 sequence from miRBase to identify pre-miRNA in the I. nil EST database. Based on comparison of the BlastN indicated sequence EST no. CJ744218 with mature and precursor sequences of miR319 from miRBase, we found the existence of I. nil miR319 precursor. We confirmed the presence of the identified sequence in I. nil using the RT-PCR reaction. An amplified DNA fragment was sequenced and analysed for the presence of sequences of mature miR319 (Fig. 2a) and secondary structure (Fig. 2b). The miR319 miRNAs are widely distributed in plants, including mosses (Arazi et al. 2005; Axtell and Bartel 2005; Axtell et al. 2007), and their precursors, which have long fold-backs with an extended upper stem above the miRNA, are also highly conserved (Palatnik et al. 2003; Li et al. 2005; Warthmann et al. 2008). The identified sequence In-pre-miR319 also creates a long hairpin structure and putative mature miRNA sequence is located in a similar place (Fig. 2b) as in precursors found in other plant species. It can therefore be concluded that the identified sequence is a precursor miR319 and encodes a mature, functional miRNA molecule.
Identification of In-pre-miR319 allowed the authors to examine the expression of InTCP4 and InMIR319 using the qRT-PCR technique. Accumulation of InTCP4 and MIR319 mRNA was examined in the vegetative and generative organs of I. nil plants (Fig. 4). High levels of InTCP4 mRNA in cotyledons accompanied lack of accumulation of In-pre-miR319; the opposite situation occurs in hypocotyls: high expression of InMIR319 causes mRNA of InTCP4 to be almost absent (Fig. 5). This finding confirms that the amount of InTCP4 transcripts is regulated by miR319, at the level of degradation of the mRNA. However, this relationship does not occur in all organs. In apexes, there are transcripts of both genes, which is probably associated with their participation in the regulation forming the leaves, as described in tomato (Ori et al. 2007). In A. thaliana, there are three genes encoding miR319 (MIR319a, -b, -c), MIR319a and MIR319b encoding identical mature miRNA and the expression patterns for the three MIR319 genes are distinct, suggesting that these genes may have largely unique developmental functions (Nag et al. 2009; Schommer et al. 2012). It can therefore be assumed that other unidentified MIR319 genes also exist in I. nil that may influence the accumulation of mRNA InTCP4 in other tissues. Identified In-pre-miR319 contains a sequence of mature miRNA319 identical to miR319a/b A. thaliana (Fig. 2a) and InMIR319 indicates a similar tissue expression pattern to MIR319a and MIR319b in A. thaliana (Nag et al. 2009). The results of Nag et al. (2009) suggest that miR319a/b might not play an important role in leaf development, but in petal and stamen development. In A. thaliana jaw-D mutants (overexpression of MIR319 gene) have slightly shorter hypocotyls than wild-type plants, whereas increased TCP activity leads to longer hypocotyls (Palatnik et al. 2003; Schommer et al. 2008; Sarvepalli and Nath 2011). In our study, we observed in hypocotyls of I. nil, high accumulation of In-pre-miR319 (Fig. 5b). These data confirm that the regulation of TCP4 mRNA level by miRNA is important for proper hypocotyl growth in various plant species.
InTCP4 is expressed also in the generative organs of mature I. nil (Fig. 5a). In addition, the transcript level of InTCP4 increases or decreases during pistil and stamen growth, respectively (Fig. 5c). The high level of InTCP4 transcript in pistils seems to be a particularly interesting fact because of the lack of reports of involvement of homologues of TCP4 in the development of this organ. TCP4 and miR319a, A. thaliana regulate the normal development of stamens and petals (Schommer et al. 2008; Nag et al. 2009). Analysis of cin mutants in Antirrhinum showed that the miRNA-regulated TCP gene, CIN, affects petal lobe development by controlling epidermal cell differentiation and growth (Crawford et al. 2004).
Apart from affecting the organ development, the TCPs have also been shown to be involved in the processes of phase change. The TCP4 mutant and jaw-D display a moderate late-flowering phenotype with an increase in leaf number compared to the wild type (Palatnik et al. 2003; Schommer et al. 2008; Sarvepalli and Nath 2011). Plants with the hyper-active form of TCP4 flower earlier (Sarvepalli and Nath 2011). The places of perception of the signal in the path of photoperiodic flowering induction are cotyledons and/or leaves (Vince-Prue and Gressel 1985; Ogawa and King 1990), so high expression of InTCP4 in cotyledons of I. nil (Fig. 5a) suggests that it may play an important function in flower induction or another process.
To investigate the possible participation of InTCP4 in the mechanism of flower induction in I. nil we investigated the effect of LD and SD conditions and factors that repress the flowering induction of I. nil on expression of InTCP4 (Fig. 6). We observed that the level of InTCP4 transcript successively decreased during both long and short night and we revealed that light positively influences the level of InTCP4 transcript accumulation. In addition, InTCP4 expression showing a diurnal cycle in the cotyledons of seedlings cultivated under both the SD and LD conditions seems to be the consequence of an endogenous rhythm. Oscillations in the level of InTCP4 mRNA are also observed in the case of IAA-treated plants (Fig. 6). The application of IAA stimulates the biosynthesis of JA, JAMe and ethylene, thus inhibiting flower induction of I. nil (Maciejewska et al. 2004; Kęsy et al. 2008, 2010, 2012; Frankowski et al. 2009). No response to exogenous IAA application, and consequently to an increase in the level of jasmonic acid, indicates that the expression of InTCP4 is not influenced by these phytohormones. This was confirmed by the results of our experiment with the induction of senescence in the cotyledon of I. nil by application of JAMe (see below). Based on the results obtained, it can be concluded that InTCP4 is not directly involved in photoperiodic flower induction of I. nil. Research in A. thaliana showed that the TCP4 gene shows daily oscillations of expression, with similar patterns as in our work (Giraud et al. 2010). Giraud et al. (2010) suggest that TCP transcription factors appear to be the central regulators in circadian clock input sites, the clock oscillator itself, and a myriad of clock output sites, directing and regulating the expression of genes.
Examination of leaf senescence revealed that jaw-D leaves had a delay in senescence, while high levels of TCP4 caused a premature onset of this process (Schommer et al. 2008). The mechanistic pathway that leads to the activation of senescence by the TCPs is currently unknown, although there might be interactions with the regulation of JA biosynthesis, as this phytohormone has been proposed to be a critical factor in senescence (Buchanan-Wollaston et al. 2003; van der Graaff et al. 2006; Schommer et al. 2008). Therefore, it was decided to examine changes in the expression of InTCP4 and InLOX2 during cotyledon senescence. Due to the very low expression of InMIR319 in cotyledon, its activity has not been investigated. Senescence was induced by darkness, JAMe, and ABA (Weaver et al. 1998; Gan 2004). The following stages of the cotyledons’ senescence process were controlled by analysis of photosynthetic parameters, such as chlorophyll content, which is typical of an ageing marker (Oh et al. 1996; Woo et al. 2001; Gan 2004; Lim et al. 2007). Furthermore, we analysed the expression of the gene associated with the function of the photosynthetic apparatus, InCAB2 encoding the chlorophyll-binding protein (Lim et al. 2007).Van der Graaff et al. (2006) showed that the CAB gene is strongly and continuously negatively regulated during the developmental and darkness-induced ageing of leaves. Based on the analysis of the level of chlorophylls and InCAB2 gene expression, we can conclude that all used factors caused ageing of cotyledons of I. nil (Fig. 7a, b). A slight decrease in the parameters in plants cultivated under control conditions (LD) (Fig. 7a, b) shows the natural cotyledons ageing due to the appearance of leaves in the 8-day-old seedlings (the second day of the experiment). In plants without access to light compared to those growing in LD we observed a strong reduction in the level of InTCP4 mRNA (Fig. 7d). The decrease in InTCP4 expression in the dark confirms the results of our experiments in LD and SD conditions and indicates the important influence of light on transcriptional activity of the gene. In the cotyledons of plants grown in LD (Fig. 6d) the expression of InTCP4 shows a similar pattern as InCAB2 (Fig. 7b), which may indicate a relationship of InTCP4 with ontogenetic senescence of cotyledons in I. nil. InTCP4 expression does not correlate with accumulation of InLOX2 mRNA, indicating a lack of direct linkage of these two genes in I. nil. InLOX2 gene transcription activity increases in the 2nd and 4th days of the experiment in plants grown in the light, and in the darkness remains at a low level, while the application of ABA and JAMe strongly promotes InLOX2 expression (Fig. 7c). Van der Graaff et al. (2006) showed a coordinated increase in AtLOX2 gene expression only during ontogenetic ageing of the leaves, and not during ageing induced by darkness. This suggests that the jasmonic pathway in leaves is specifically activated by development cues. Furthermore, the A. thaliana LOX2 responds positively to wounding and JAMe (Bell et al. 1995; Jensen et al. 2002). Our results confirm the data by van der Graaff et al. (2006) that activation of the LOX2 gene is associated with developmental senescence rather than induced by darkness. In addition, the results confirm the presence of a positive feedback loop, namely, that the JAMe stimulates the expression of jasmonic biosynthesis genes (Jensen et al. 2002; Acosta and Farmer 2010).
To conclude, the results of this study do not provide a clear answer whether InTCP4 is involved in ageing cotyledons of I. nil, but it did not exclude the possibility. Perhaps it is involved in the regulation of this process, not by induction of expression of genes involved in jasmonic acid biosynthesis, but by other genes which were not studied. Similar results were obtained by Sarvepalli and Nath (2011). They observed that the expression level of AtLOX2 was significantly reduced in tcp4-1 mutant plants, but it was not up-regulated in TCP4:VP16-C plants (hyper-active TCP4). These data raise the possibility that TCP4 mediates senescence by an LOX2-independent pathway (Sarvepalli and Nath 2011).
Based on the results obtained in our research, it can be concluded that both light (clock) and miR319 control the level of InTCP4 transcript. The main function of InMIR319 seems to be the regulation of InTCP4 organ localization. Our research suggests that InTCP4 and InMIR319 participate in the control of such processes as do their homologues in other plant species, i.e., the development of cotyledons, leaves and flower elements.
Author contribution
Paulina Glazińska designed and carried out the experiments, analysed the data and wrote the manuscript. Emilia Wilmowicz and Waldemar Wojciechowski carried out the experiments. Kamil Frankowski was responsible for the preparation of figures. Jan Kopcewicz helped in preparing the manuscript.
References
Acosta IF, Farmer EE (2010) Jasmonates. In: Arabidopsis Book 8:e0129. doi:10.1199/tab.0129
Aggarwal P, Das Gupta M, Joseph AP, Chatterjee N, Srinivasan N, Nath U (2010) Identification of specific DNA binding residues in the TCP family of transcription factors in Arabidopsis. Plant Cell 22:1174–1189. doi:10.1105/tpc.109.066647
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Arazi T, Talmor-Neiman M, Stav R, Riese M, Huijser P, Baulcombe DC (2005) Cloning and characterization of micro-RNAs from moss. Plant J 43:837–848. doi:10.1111/j.1365-313X.2005.02499.x
Aukerman MJ, Sakai H (2003) Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-Like target genes. Plant Cell 15:2730–2741. doi:10.1105/tpc.016238
Axtell MJ, Bartel DP (2005) Antiquity of microRNAs and their targets in land plants. Plant Cell 17:1658–1673. doi:10.1105/tpc.105.032185
Axtell MJ, Snyder JA, Bartel DP (2007) Common functions for diverse small RNAs of land plants. Plant Cell 19:1750–1769. doi:10.1105/tpc.107.051706
Baker CC, Sieber P, Wellmer F, Meyerowitz EM (2005) The early extra petals1 mutant uncovers a role for microRNA miR164c in regulating petal number in Arabidopsis. Curr Biol 15:303–315. doi:10.1016/j.cub.2005.02.017
Bell E, Creelman RA, Mullet JE (1995) A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc Natl Acad Sci USA 92:8675–8679
Bonnet E, Van de Peer Y, Rouzé P (2006) The small RNA world of plants. New Phytol 171:451–468. doi:10.1111/j.1469-8137.2006.01806.x
Buchanan-Wollaston V, Earl S, Harrison E, Mathas E, Navabpour S, Page T, Pink D (2003) The molecular analysis of leaf senescence—a genomics approach. Plant Biotechnol J 1:3–22
Crawford BC, Nath U, Carpenter R, Coen ES (2004) CINCINNATA controls both cell differentiation and growth in petal lobes and leaves of Antirrhinum. Plant Physiol 135:244–253. doi:10.1104/pp.103.036368
Cubas P, Lauter N, Doebley J, Coen E (1999) The TCP domain: a motif found in proteins regulating plant growth and development. Plant J 18:215–222. doi:10.1046/j.1365-313X.1999.00444.x
Frankowski K, Kesy J, Wojciechowski W, Kopcewicz J (2009) Light- and IAA-regulated ACC synthase gene (PnACS) from Pharbitis nil and its possible role in IAA-mediated flower inhibition. J Plant Physiol 166:192–202. doi:10.1016/j.jplph.2008.02.013
Gan S (2004) The hormonal regulation of senescence. In: Davies PJ (ed) Plant hormones: biosynthesis, signal transduction and action!. Kluwer Academic Publishers, Dordrecht, pp 561–581
Giraud E, Ng S, Carrie C, Duncan O, Low J, Lee CP, Aken OV, Millar AH, Murcha M, Whelan J (2010) TCP transcription factors link the regulation of genes encoding mitochondrial proteins with the circadian clock in Arabidopsis thaliana. Plant Cell 22:3921–3934. doi:10.1105/tpc.110.074518
Glazińska P, Zienkiewicz A, Wojciechowski W, Kopcewicz J (2009) The putative miR172 target gene InAPETALA2-like is involved in the photoperiodic flower induction of Ipomoea nil. J Plant Physiol 166:1801–1813. doi:10.1016/j.jplph.2009.05.011
Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ (2008) miRBase: tools for microRNA genomics. Nucleic Acids Res 36:D154–D158. doi:10.1093/nar/gkm952
Hayama R, Coupland G (2004) The molecular basis of diversity in the photoperiodic flowering responses of Arabidopsis and Rice. Plant Physiol 135:677–684. doi:10.1105/tpc.107.052480
Hayama R, Agashe B, Luley E, King R, Coupland G (2007) A circadian rhythm set by dusk determines the expression of FT homologs and the short-day photoperiodic flowering response in Pharbitis. Plant Cell 19:2988–3000. doi:10.1105/tpc.107.052480
Imaizumi T, Kay SA (2006) Photoperiodic control of flowering: not only by coincidence. Trends Plant Sci 11:550–558. doi:0.1016/j.tplants.2006.09.004
Izawa T, Oikawa T, Sugiyama N, Tanisaka T, Yano M, Shimamoto K (2002) Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Dev 16:2006–2020. doi:10.1101/gad.999202
Jensen AB, Raventos D, Mundy J (2002) Fusion genetic analysis of jasmonate-signalling mutants in Arabidopsis. Plant J 29:595–606. doi:10.1046/j.0960-7412.2001.01241.x
Jones- Rhoades MW, Bartel DP, Bartel B (2006) MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol 57:49–53. doi:10.1146/annurev.arplant.57.032905.105218
Kasschau KD, Xie Z, Allen E, Llave C, Chapman EJ, Krizan KA, Carrington JC (2003) P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev Cell 4:205–217. doi:10.1016/j.devcel.2011.02.003
Kęsy J, Maciejewska B, Sowa M, Szumiak M, Kawałowski K, Borzuchowska M, Kopcewicz J (2008) Ethylene and IAA interactions in the inhibition of photoperiodic flower induction of Pharbitis nil. Plant Growth Regul 55:43–50. doi:10.1007/s10725-008-9256-9
Kęsy J, Frankowski K, Wilmowicz E, Glazińska P, Wojciechowski W, Kopcewicz J (2010) The possible role of PnACS2 in IAA-mediated flower inhibition in Pharbitis nil. Plant Growth Regul 61:1–10. doi:10.1007/s10725-010-9443-3
Kęsy J, Wilmowicz E, Maciejewska B, Frankowski K, Glazińska P, Kopcewicz J (2012) Independent effects of jasmonates and ethylene on inhibition of Pharbitis nil flowering. Acta Physiol Plantarum 33:1211–1216. doi:10.1007/s11738-010-0649-9
Kidner CA (2010) The many roles of small RNAs in leaf development. J Genet Genomics 37:13–21. doi:10.1016/S1673-8527(09)60021-7
Kosugi S, Ohashi Y (2002) DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J 30:337–348. doi:10.1046/j.1365-313X.2002.01294.x
Laufs P, Peaucelle A, Morin H, Traas J (2004) MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development 131:4311–4322. doi:10.1242/dev.01320
Lauter N, Kampani A, Carlson S, Goebel M, Moose SP (2005) microRNA172 down-regulates glossy15 to promote vegetative chase change in maize. Proc Natl Acad Sci USA 102:9412–9417. doi:10.1073/pnas.0503927102
Li Y, Li W, Jin YX (2005) Computational identification of novel family members of microRNA genes in Arabidopsis thaliana and Oryza sativa. Acta Biochim Biophys Sin (Shanghai) 37:75–87. doi:10.1093/abbs/37.2.75
Lim PO, Kim HJ, Nam HG (2007) Leaf senescence. Annu Rev Plant Biol 58:115–136. doi:10.1146/annurev.arplant.57.032905.105316
Liu PP, Montgomery TA, Fahlgren N, Kasschau KD, Nonogaki H, Carrington JC (2007) Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J 52:133–146. doi:10.1111/j.1365-313X.2007.03218.x
Maciejewska BD, Kęsy J, Zielinska M, Kopcewicz J (2004) Jasmonates inhibit flowering in short-day plant Pharbitis nil. Plant Growth Regul 43:1–8. doi:10.1023/B:GROW.0000038241.00771.bd
Mallory AC, Dugas DV, Bartel DP, Bartel B (2004) MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr Biol 14:1035–1046. doi:10.1016/j.cub.2004.06.022
Mallory AC, Bartel DP, Bartel B (2005) MicroRNA- directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 17:1360–1375. doi:10.1105/tpc.105.031716
Martin-Trillo M, Cubas P (2010) TCP genes: a family snapshot ten years later. Trends Plant Sci 15:31–39. doi:10.1016/j.tplants.2009.11.003
Nag A, King S, Jack T (2009) miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc Natl Acad Sci USA 106:22534–22539. doi:10.1073/pnas.0908718106
Navaud O, Dabos P, Carnus E, Tremousaygue D, Hervé C (2007) TCP transcription factors predate the emergence of land plants. J Mol Evolution 65:23–33. doi:10.1007/s00239-006-0174-z
Ogawa Y, King RW (1990) The inhibition of flowering by non-induced cotyledons of Pharbitis nil. Plant Cell Physiol 31:129–135
Oh SA, Lee SY, Chung IK, Lee CH, Nam HG (1996) A senescence-associated gene of Arabidopsis thaliana is distinctively regulated during natural and artificially induced leaf senescence. Plant Mol Biol 30:739–754
Ori N, Cohen AR, Etzioni A, Brand A, Yanai O, Shleizer S, Menda N, Amsellem Z, Efroni I, Pekker I, Alvarez JP, Blum E, Zamir D, Eshed Y (2007) Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat Genet 39:787–791. doi:10.1038/ng2036
Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D (2003) Control of leaf morphogenesis by microRNAs. Nature 425:257–263. doi:10.1038/nature01958
Parthier B (1990) Jasmonates: hormonal regulators of stress factors in leaf senescence. J Plant Growth Regul 9:445–454. doi:10.1007/BF02041942
Pulido A, Laufs P (2010) Co-ordination of developmental processes by small RNAs during leaf development. J Exp Bot 61:1277–1291. doi:10.1093/jxb/erp397
Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP (2002) MicroRNAs in plants. Genes Dev 16:1616–1626. doi:10.1101/gad.1004402
Rubio-Somoza I, Weigel D (2011) MicroRNA networks and developmental plasticity in plants. Trends Plant Sci 16:258–264. doi:10.1016/j.tplants.2011.03.001
Sarvepalli K, Nath U (2011) Hyper-activation of the TCP4 transcription factor in Arabidopsis thaliana accelerates multiple aspects of plant maturation. Plant J 67:595–607. doi:10.1111/j.1365-313X.2011.04616.x
Schommer C, Palatnik JF, Aggarwal P, Chetelat A, Cubas P, Farmer EE, Nath U, Weigel D (2008) Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol 6:1991–2001. doi:10.1371/journal.pbio.0060230
Schommer C, Bresso EG, Spinelli SV, Palatnik JF (2012) Role of microRNA miR319 in plant development. In: Sunkar R (ed) MicroRNAs in plant development and stress responses. Signaling and communication in plants, vol 15. Springer, Heidelberg, pp 29–47
Sieber P, Wellmer F, Gheyselinck J, Riechmann JL, Meyerowitz EM (2007) Redundancy and specialization among plant microRNAs: role of the MIR164 family in developmental robustness. Development 134:1051–1060. doi:10.1242/dev.02817
Thomas B, Vince-Prue D (1997) Photoperiodism in plants, 2nd edn. Academic Press Inc, San Diego, p 428
Turner JG, Ellis C, Devoto A (2002) The jasmonate signal pathway. Plant Cell (Suppl.): S153–S164. doi:10.1105/tpc.000679
Ueda J, Kato J (1980) Identification of a senescence-promoting substance from wormwood (Artemisia absinthum L). Plant Physiol 66:246–249
van der Graaff E, Schwacke R, Schneider A, Desimone M, Flugge UI, Kunze R (2006) Transcription analysis of arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol 141:776–792. doi:10.1104/pp.106.079293
Vince-Prue D, Gressel J (1985) Pharbitis nil. In: Halevy AH (ed) Handbook of flowering IV. CRC Press Inc, Boca Raton, pp 47–81
Voinnet O (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136:669–687. doi:10.1016/j.cell.2009.01.046
Wang JW, Wang LJ, Mao YB, Cai WJ, Xue HW, Chen XY (2005) Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant Cell 17:2204–2216. doi:10.1105/tpc.105.033076
Warthmann N, Das S, Lanz C, Weigel D (2008) Comparative analysis of the MIR319a microRNA locus in Arabidopsis and related Brassicaceae. Mol Biol Evol 25:892–902. doi:10.1093/molbev/msn029
Weaver LM, Gan S, Quirino B, Amasino RM (1998) A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Mol Biol 37:455–469. doi:10.1023/A:1005934428906
Woo HR, Chung KM, Park JH, Oh SA, Ahn T, Hong SH, Jang SK, Nam HG (2001) ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell 13:1779–1790. doi:10.1105/TPC.010061
Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS (2009) The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138:750–759. doi:10.1016/j.cell.2009.06.031
Yoshida S (2003) Molecular regulation of leaf senescence. Curr Opin Plant Biol 6:79–84. doi:10.1016/S1369-5266(02)00009-2
Zhang B, Pan X, Cannon CH, Cobb GP, Anderson TA (2006) Conservation and divergence of plant microRNA genes. Plant J 46:243–259. doi:10.1111/j.1365-313X.2006.02697.x
Zhang B, Wang Q, Pan X (2007) MicroRNAs and their regulatory roles in animals and plants. J Cell Physiol 210:279–289. doi:10.1002/jcp.20869
Acknowledgments
The work was supported by the Polish Ministry of Science and Higher Education grant N303 025 321637 and Nicolaus Copernicus University in Torun, Poland grant 538-B.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J.-H. Liu.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Glazińska, P., Wilmowicz, E., Wojciechowski, W. et al. Impact of InMIR319 and light on the expression of InTCP4 gene involved in the development of Ipomoea nil plants. Acta Physiol Plant 36, 29–43 (2014). https://doi.org/10.1007/s11738-013-1384-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11738-013-1384-9