Abstract
The quantitative analysis of andrographolides in Andrographis paniculata plant materials is essential for pharmaceutical factories. This analysis cannot be done for all samples due to the conventional process using the extraction and HPLC methods requires a long analysis time and sample destruction. Therefore, near-infrared spectroscopy (NIRS) was employed to classify the class of A. paniculata and to determine the content of two active ingredients, andrographolide (AP1) and dehydroandrographolide (AP3) in A. paniculata, rapidly and non-destructively. One hundred twenty dried powder samples were obtained from aerial parts, branches, leaves, and branches mixed with leaves. The NIR absorption scans were collected from a broad spectral region (1000–2500 nm). Then, the scanned samples were extracted and analyzed for their AP1 and AP3 contents using an HPLC reference method. The success classification model based on AP1 level was developed using the second derivative pretreated NIR spectra of the entire wavelength region using the Partial Least Squares-Discriminant Analysis (PLS-DA) method. The NIR calibration models were developed and tested for quantitative analysis with 50 independent samples. The models were identified for the analysis of the AP1 content with excellent performance (correlation coefficient (R) = 0.98; standard error of validation (SEV) = 0.24%) and for the analysis of the AP3 content at a good level of efficiency (R = 0.93; SEV = 0.15%). This study showed that NIR spectroscopic method offers rapid analysis for the selection of A. paniculata that meets the requirement in bioactive amount.
Graphic Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Recently, Andrographis sp. (Andrographis paniculata (Burm. F.) Wall. Ex Nees), also known as “Xiyanping” in Chinese, is one of the target medicinal plants that are under testing for the anti a novel coronavirus (SARS-CoV-2) caused the coronavirus disease 2019 (COVID-19) in Thailand, China, and other countries (Sa-ngiamsuntorn et al. 2021; Enmozhi et al. 2020; Murugan et al. 2020; Shi et al. 2020; Cai et al. 2020). In general, A. paniculata is indicated for the relief of the common cold associated with nasal congestion, such as a sore throat, muscle aches, mucus, and stomach ache (Pholphana et al. 2004) and is more active as an anti-inflammatory (Shen et al. 2000) and related immunity (Puri et al. 1993; Calabrese et al. 2000; Chowdhury et al. 2012). Furthermore, andrographolide compound in A. paniculata was explored and reported with respect to the activity of broad-spectrum antiviral properties (Gupta et al. 2017). In 2020, Enmozhi et al. (Enmozhi et al. 2020) evaluated the andrographolide from A. paniculata as a potential inhibitor of the main protease of SARS-COV-2 (Mpro) through in silico studies such as molecular docking, target analysis, toxicity prediction, and the absorption, distribution, metabolism, excretion (ADME) prediction. Moreover, Cai et al. (Cai et al. 2020) reported that A. paniculata could reduce inflammation in COVID-19 patients and improve symptoms such as cough, fever, and rales in the lungs. It is recommended for treating patients in the progressive stage of COVID-19 (critical case) in the National Health Council (NHC) guidelines.
Therefore, the plant materials of A. paniculata should be confirmed to ensure the active content of andrographolide within a level of acceptance. Thai Herbal Pharmacopoeia (Department of Medical Sciences, Ministry of Public Health 2019) defined the positive factors for a powder of the aerial part of A. paniculata herb plant, with the amount of A. paniculata not less than 1.0%(w/w). However, there are significant differences in the active compound depending on the specific part of the herb plant collected, geographic origin, season, and harvest-time (Hossain et al. 2014). It has been reported that the greatest amount of bioactive compound from A. paniculata increased at 110 days postharvest and was lower at 130 days after pre-blossom of the plant (Sharma and Sharma 2013). There appear to be problems with A. paniculata, in specific situations, regarding the raw materials for the herb business and their regular supply and quality that are essential for a commercial enterprise. The purchase price of raw materials is usually settled based on the physical appearance involving piece form, color and contaminants, trust between the buyer and seller regarding the time of harvest, and that there are substantial bioactive compounds. However, it is possible for quality control in collection and processing to be variable, resulting in lost value for downstream products. The quantitative analysis of bioactive A. paniculata components is generally based on the extraction and analysis to estimate the amount of diterpene lactone using various methods such as TLC (Rajani et al. 2000), LC (Jain et al. 2000), and HPLC (Chen et al. 2007; Xu et al. 2008; Sharma et al. 2012). However, there are limitations in the analysis, and because these processes require considerable time, samples can be damaged through using the chemical substance and several types of scientific equipment. Thus, the high cost of analysis and the complex procedures require experienced analysts. The current research aimed to find possible opportunities by focusing on quality control upstream in the process and specifically on the raw material, using NIR spectroscopy to test for the amount of active ingredients in the A. paniculata samples, and thus solve these problems.
The application of NIR to analyze the bioactive compound of herbs has started to spread worldwide, especially in Asian countries such as China, Korea, Japan, and Thailand. There has been researched into both quantitative analysis of the active ingredients and quality analysis to collect and identify genuine herbs to command high prices based on the level of potential substrates categorized by geographical origin. In our previous studies, we reported the application of moving window partial least squares regression (MWPLSR) to the quantitative determination of total curcuminoids in turmeric rhizome by NIR spectroscopy (Kasemsumran et al. 2010). The second derivative pretreated NIR spectral range of 2040–2486 nm was selected to build the model for total curcuminoids prediction in turmeric powder sample, in which the obtained statistical values were the standard error of prediction (SEP) of 1.003% w/w and the ratio of prediction to deviation (RPD) of 4.857. We extended our study for NIR applications to a pharmaceutical analysis by reporting the achievement of NIR assaying curcumin in the 170-capsule of turmeric herbal medicines (Kasemsumran et al. 2014). Some works including our studies are summarized in Table 1 (Kasemsumran et al. 2010, 2014, 2017; Ren and Chen 1999; Schulz et al. 1999; Luypaert et al. 2003; Chen et al. 2009; Lee et al. 2014; McGoverin et al. 2010; Chan et al. 2007; Wang et al. 2007; Lim et al. 2012; Tahir et al. 2020; Liu et al. 2018; Tanaka et al. 2008; Kim et al. 2014). In addition to those research works, there has been a study to analyze the amount of andrographolides in Chinese herb samples of A. paniculata that developed a good calibration model with good efficiency and accurate prediction (Lai et al. 2018). However, the parts of A. paniculata plants didn’t take into account in the study.
The objective of this research is to investigate the efficiency of NIR technology aiming at the classification for a selection process of A. paniculata with high bioactive compound and quantification of andrographolide (AP1) and dehydroandrographolide (AP3) in A. paniculata. The 170 samples with different sources and plant parts were achieved in this study since the quantity of bioactive compounds varies in different plant parts of A. paniculata. The developed process was achieved using the long-wavelength NIR region of 1000–2500 nm to measure the diffuse reflectance spectra of A. paniculata and partial least squares-discriminant analysis (PLS-DA) combined with partial least squares regression (PLSR) to construct the discriminant model and quantitative model, respectively.
Partial least squares regression (PLSR)
PLSR is usually employed in many applications for quantitative analysis (Kasemsumran et al. 2014; Schulz et al. 1999; Luypaert et al. 2003; Chen et al. 2009; McGoverin et al. 2010; Chan et al. 2007; Wang et al. 2007; Lim et al. 2012; Tanaka et al. 2008; Kim et al. 2014). It is a very powerful method, in which the y-variable (reference value) is taken to account by balancing the X- and y-information, then, the factors as linear combinations of the original spectral data (x-values) are constructed and employs only these factors in the regression equation. PLS regression aims to reduce the quantity of spectral data, only the most relevant part of the x-variation is used in a regression for predicting y. Thus, an efficient PLS calibration model can be obtained.
Partial least squares-discriminant analysis (PLS-DA)
PLS-DA is based on the PLSR algorithm. It is suitable for the classification of high dimension NIR spectral data for diverse purposes (Kasemsumran et al. 2017). It optimizes the fitting and prediction y to {0/1}-coded membership indicating variables in the development of factor number.
For quality control of herbal samples by using NIR spectroscopy, the support of advanced chemometric method is vital and helpful to achieve an efficient calibration model of the analyte.
Experimental
Materials
Figure 1 shows the form of a typical A. paniculata plant. For the current study, 170 samples of A. paniculata of know origin were used. Samples were collected over two years from four parts of each plant: aerial parts, stems, leaves, and stems mixed with leaves (Fig. 2) from farmland and herbal drugstores in ten provinces of Thailand namely, Bangkok, Nakhon Pathom, Nonthaburi, Prajeenburi, Phetchaburi, Phetchabun, Mahasarakham, Suphanburi, and Sa Kaeo. Fresh samples were heated at 50 °C for 10 h and then, after this was ground in a mechanical grinder (Cyclotec, model 1093 FOSS, Hillerod, Denmark) and passed through a 1 mm sieve.
FT-NIR spectral acquisition
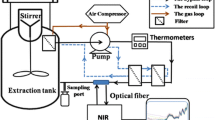
1.00 ± 0.02 g of each powdered sample was kept in a separate glass vial and tightly pressed using stainless-steel seal equipment. All samples were collected with approximately the same depth of 1.2 cm above the vial bottom. The sample vial was placed on the vial measuring tray for NIR measurement as shown in Fig. 3, and data were collected using a Fourier Transform—Near-Infrared Spectrophotometer (FT-NIR) (NIR-Flex Solid, Buchi, Switzerland) in the scan range of 10,000–4000 cm−1. Triplicate measurements were made for each sample at a spectral resolution of 8 cm−1 and 128 times per scan. It is noted that the units of reflectance and wavenumber were converted to absorbance and wavelength (1000–2500 nm) prior to data analysis for the easy NIR band assignment.
Bioactive compound extraction and quantification using HPLC
The powders, as scanned by NIR, were subjected to methanol extraction (AR Grade; ACI Labscan), in which the sample was placed in a cellulose thimble (30 mm × 100 mm) and extracted using methanol of 150 ml for 210 min in a soxhlet system. The extract was then concentrated using a rotary evaporator (Model R-210/215; Buchi; Denmark) and completely dried in an oven. Each concentrated extract was diluted by methanol (HPLC grade; ACI Labscan) to adjust the volume. The resultant liquid was passed through a syringe filter and collected in a tinted vial for HPLC analysis.
Bioactive compound quantification was done using HPLC according to Xu et al. (2008) by injecting a 10 μl sample into a column (Inert Sustain C18, 4.6 mm ID × 150 mm 5 μm; HPLC Shimadzu model LC-20A) with methanol: water (55:45) as the mobile phase at a flow rate of 1.0 ml/min at 30 °C with a UV detector at 223 nm. The standard curve was compiled based on the average from duplicates of standard andrographolide (purity ≥ 98%; Sigma-Aldrich) and dehydroandrographolide (purity ≥ 98; Sigma-Aldrich). Five calibrators of standard were freshly prepared by diluting the stock solutions with mobile phase in appropriate quantities. The calibration range was 10–300 μg/ml for both andrographolide (AP1) and dehydroandrographolide (AP3). The peak-area versus AP1 and AP3 concentrations in the calibration range resulted in the regression equation, r = 0.9999. Moisture analysis in herb was also done for all powder samples, and the results were employed for concentration calculation on a dry basis.
Statistical analysis of mean quantity from different parts of A. paniculata
Data of the bioactive quantities from AP1 and AP3 based on HPLC from the 120 collected samples were processed and then subjected to one-way analysis of variance, and sample means were compared Duncan’s new multiple range test at α = 0.05.
Calibration model development for discrimination and quantitative analysis of AP1 and AP3 content in A. paniculata using NIR spectral data
The NIR model was constructed using Unscrambler version 9.8 (CAMO AS; Trondheim, Norway) with three modes of the spectrum: (1) uncorrected spectrum; (2) corrected spectrum using multiplicative scattering correction (MSC); and (3) corrected spectrum using second derivatives (2D) based on Savitzky-Golay model (polynomial order = 2, number of smoothing points = 5), while applied with PLS technique using PLS1. All 170-sample data were divided into 120 calibration sets with the remaining 50 samples in the validation set. Full cross-validation was used to find the optimum number of PLS factors (F) with the lowest standard error of cross-validation (SECV) for the models used for the discrimination and quantitative determination.
The selection of A. paniculata herb plant by the amount of AP1 as a major bioactive compound was required not less than 1.0%. For the rapid selection, the classification model was built using PLS-DA. The calculation involved giving values of “0” and “1” to the NIR spectra of samples having a content of AP1 lower than 1.0% (out of specification) and that the high content from 1.0% (in the specification), respectively. The classification PLS model was built in a regression for predicting y. A sample with the predicted result lower than 0.5 was classified as out of specification (OUT), else the predicted result greater than equal 0.5 was classified as meet a requirement (PASS-1). Furthermore, individual PLS calibration models for the quantitative determination of actual AP1 and AP3 contents in A. paniculata were developed, respectively.
Results and discussion
Quantitative analysis of AP1 and AP3 in A. paniculata using HPLC
Figure 4 shows the chromatogram of an extracted sample having the highest amount of AP1 and less AP3, with peak retention times of 5.31 min and 14.39 min, respectively. Therefore, the major active compound found in A. paniculata plants was AP1.
From the 120-sample calibration set, Table 2 shows the different amounts of both AP1 and AP3 extracted from the different parts of A. paniculata plants. The leaves contained the highest proportion of AP1. The aerial and leaf parts contained the highest proportion of AP3, without any significant difference between the AP3 levels from these two parts. Therefore, the leaves are the most important part to be used as raw material for extraction to acquire bioactive compounds since they contained the highest proportions of both AP1 and AP3. In addition, the results of the study indicated that commercial samples used in this study might be harvested during the period of vegetative, but not more than 50% of the flowering (Chen et al. 2007).
All samples (in both the calibration and validation sets) contained AP1 and AP3 in the range 0.151–3.608% and 0.572–1.990% by dry weight, respectively. Table 3 shows the distribution of quantities for the samples in the calibration and validation sets. The histogram in Fig. 5 indicates that amounts of AP1 and AP3 appeared to be normally distributed for both the calibration and validation sets. In fact, the normal curve line for the AP1 content differed more from a standard curve than for AP3. This indicated that each part contained many different quantities of AP1 with a higher resultant standard deviation (SD) of 0.900–0.902% dry weight than for AP3. The AP3 content appeared to be high curve (low flatness) for a normal distribution. Therefore, the SD was lower (0.336–0.338% dry weight), indicating that the selection of the raw material can be more rapid. Considering only the quantity of AP1, a selection of leaves would be best to maximize production. However, after grinding, it would be difficult to distinguish just this part of the material. Based on these results, it was necessary to develop a method to quantify the contents by applying a NIR spectroscopy technique.
NIR spectral data of A. paniculata powder
The standard normal variate (SNV) of three mean original NIR spectra in the 1000–2500 nm region of A. paniculata powder sample (dot line), standard AP1 (solid line), and standard AP3 (dash line) is illustrated in Fig. 6. It can be seen the informative bands due to the NIR active functional group (Fig. 4) of AP1 and AP3 in the NIR spectrum of A. paniculata sample as follows; two bands at 1180 and 1380 nm are the second-overtone of C-H alkene, and C-H methyl-alicyclic hydrocarbon, individually (Workman and JrL 2007). Two broad bands around 1430 and 1940 nm are largely due to the combination of O–H symmetric and antisymmetric stretching modes of water and the combination mode of the O–H stretching and deformation vibrations of water, respectively (Maeda et al. 1995). The water bands come from the residual moisture in the powder sample. A small peak around 1630 nm raises by the 1st overtones of C–H alkene (ethylidene). One broad band around 1660–1800 nm is assigned to the C–H and O–H of alicyclic hydrocarbon, and the C–H methylene. An individual peak at 1940 (O–H bending) and 1950 nm (O–H and C–H combination) was found in the NIR spectrum of the AP3 and AP1 standards, respectively. However, these two peaks did not appear in the NIR spectrum of A. paniculata sample because they were covered with a broad NIR band due to water. Those bands in 2060–2200 and 2270–2500 nm were associated with the combination band of C–H alkene and O–H, and the combination band of C–H stretching, C–H2 deformation of alicyclic hydrocarbon of AP1 and AP3 standards (Workman and JrL 2007).
Figure 7A shows the NIR spectra of all powder samples based on absorbance by second derivatives (2D) using the Savitzky-Golay model (polynomial order = 2, number of smoothing points = 5). Using the 2D, it was possible to separate the overlapping spectra to understand better the relation between the light absorption and structural molecules as previously described. Figure 7B illustrates the SNV of two mean NIR spectra in the region of 1000–2500 nm of A. paniculata powder contained low and high AP1 content between 1.0%. It can be seen that the intensity of the water bands keeps constant with that comparison of two mean spectra having different in the content of AP1 in the samples (Fig. 7B). On the other hand, the intensity of the band in the 2060–2400 nm wavelength region changes with a change in AP1 content in samples (Fig. 7B). This region is relatively rich in the combination bands of C–H and O–H and C–H alicyclic hydrocarbon (Workman and JrL 2007). Nevertheless, those mean NIR spectra are very similar, and it is not easy to classify them. Therefore, the PLS-DA method was applied in this study to classify between samples with high (≥ 1.0%) and low (< 1.0%) amounts of AP1 in A. paniculata samples.
Results of NIR calibration model for discrimination analysis of A. paniculata based on AP1 content
The classification models were developed using the different pretreatment spectra. The classification performances of PLS-DA regression models were compared, and their statistical results were reported in Table 4. The results show that the PLS-DA method yields success classification results in both none and pretreatment methods. The %CC value obtained from the PLS-DA method was 100% (Figure S1). The standard errors of calibration (SEC) and validation (SEV) are the important factors that express the classification performance in this study. The best PLS-DA model for the selection process of A. paniculata based on AP1 content was developed using 2D spectra in the whole region of 1000–2500 nm with a PLS factor of 6. It gives the best classification performance of 100% (CC) with the lowest SEC and SEV values. It appears that the 2D pretreatment can enhance any spectral differences in the whole region where bands due to any differences in the AP1 content appear. Figure 8 is a plot between the given actual value (X-axis) for each class and the PLS-DA predicted value (Y-axis) for validation samples by the best PLS-DA model.
Results of NIR calibration model for determination of AP1 and AP3 contents
Table 5 shows the calibration statistic model for quantitative analysis of AP1 and AP3 in A. paniculata based on the data collected from the entire range of NIR absorption spectra. The best NIR calibration model for the prediction of AP1 content was a calibration generated from the 2D spectra with the factor of 7. Due to the power of 2D pretreatment, one can enhance the spectral differences that are related to vary of AP1 content. It provided the standard error of calibration (SEC) at its lowest (0.191%) with the highest correlation coefficient (R = 0.977). The test results from the full cross-validation had the lowest standard error of cross-validation (SECV = 0.248%), while testing with the independent sample set had a low standard error of validation (SEV) of 0.238%.
Using the calibration dataset, the best result for determining the AP3 content used the NIR spectra without modification at the PLS factor of 11, with the lowest SEC of 0.123% and the highest correlation coefficient of R = 0.931. The test results from the full cross-validation had the lowest SECV of 0.144%, while the test results from the independent sample set also had a low SEV at 0.152%. The spectral pretreatment was unnecessary in model development for the prediction of AP3 content, perhaps, it is due to the amount, and standard deviation of AP3 in the samples is lower than that of AP1 (Table 3). The spectral optimization by those pretreatment methods in the entire wavelength region may yield positive effects on the AP1 compound because of its highest quantities and variation.
Figure 9A shows the scatter plot of the predicted AP1 contents from the best part of the NIR calibration and the data based on absorbance values with 2D on the y-axis and based on HPLC on the x-axis. By the way, Fig. 9B shows the scatter plot between the results of predicting the AP3 contents from the best NIR calibration equation when calculated from the whole range of non-pretreated NIR data (y-axis) against the actual content analyzed by HPLC (x-axis). Both of them revealed a good straight-line relationship.
Conclusions
The modeling results obtained in this study have demonstrated that NIR spectroscopy is promising to discover the target A. paniculata with high AP1 content and determine the contents of AP1 and AP3 in A. paniculata for the material selection process. PLS-DA analysis using an entire wavelength region of 1000–2500 nm and pretreated spectra with the 2D method succeed in classifying between A. paniculata materials in the specification (high AP1 of ≥ 1%) and those out of specification. Thus, an efficient PLS-DA model was obtained with 100%CC for the validation set. Furthermore, excellent PLS models for the determination of AP1 and AP3 contents in A. paniculata were developed. The most selective PLS calibration model for the quantitative determination of AP1 content in A. paniculata built by using 2D pretreated spectra over the whole region of 1000–2500 nm yielded prediction results with a low error of 0.24%. For the quantitative analysis of AP3 content in A. paniculata, PLS calibration developed using original spectra in the entire regions yielded good results with an error limit of 0.15%. Additionally, the leaf parts of A. paniculata produced the finest possible products due to its highest content of bioactive compounds. Therefore, NIR technology is a fabulous method that offers rapid analysis without extraction and without causing damage to samples that can facilitate the quality control of A. paniculata raw materials in pharmaceutical industry. This means that the quality of raw materials can be controlled for all samples. Consequently, the risk of substandard herbal products is possible to decrease.
References
Cai N, Li YP, Zhou GJ, Xin C, Xie J, Zhou HH (2020) Theoretical basis and effect characteristics of andrographolide against COVID-19 China. Chin Tradit Herb Drug 51(05):1159–66
Calabrese C, Berman SH, Babish JG, Ma X, Shinto L, Dorr M, Wells K, Wenner CA, Standish LJ (2000) A phase I trial of andrographolide in HIV positive patients and normal volunteers. Phytother Res 14:333–338
Chan CO, Chu CC, Mok DKW, Chau FT (2007) Analysis of berberine and total alkaloid content in Cortex phellodendri by near infrared spectroscopy (NIRS) compared with high-performance liquid chromatography coupled with ultra-visible spectrometric detection. Anal Chim Acta 592:121–131
Chen L, Jin H, Ding L, Zhang H, Wang X, Wang Z, Li J, Qu C, Wang Y, Zhang H (2007) On-line coupling of dynamic microwave-assisted extraction with high-performance liquid chromatography for determination of andrographolide and dehydroandrographolide in Andrographis paniculata Nees. J Chromatogr A 1140(1–2):71–77
Chen Q, Zhao J, Chaitep S, Guo Z (2009) Simultaneous analysis of main catechins contents in green tea (Camellia sinensis (L.)) by Fourier transform near infrared reflectance (FT-NIR) spectroscopy. Food Chem 113(4):1272–1277
Chowdhury A, Biswas SK, Raihan SZ, Das J, Paul S (2012) Pharmacological potentials of Andrographis paniculata: an overview. Int J Pharmacol 8(1):6–9
Department of Medical Sciences, Ministry of Public Health (2019) Fa Tha Lai. Thai Herbal Pharmacopoeia. Prachachon Co., Ltd, Thailand, pp 112–121
Enmozhi SK, Raja K, Sebastine I, Joseph J (2020) Andrographolide as a potential inhibitor of SARS-CoV-2 main protease: an in silico approach. J Biomol Struct Dyn 27:1–7
Gupta S, Mishra KP, Ganju L (2017) Broad-spectrum antiviral properties of andrographolide. Arch Virol 162:611–623
Hossain S, Urbi Z, Sule A, Rahman KMH (2014) Andrographis paniculata (Burm. f.) Wall. ex Nees: a review of ethnobotany, phytochemistry, and pharmacology. Sci World J Article ID 274905:1–28
Jain DC, Gupta MM, Saxean S, Kumar S (2000) LC analysis of hepatoprotective diterpenoids from Andrographis paniculata. J Pharm Biomed Anal 22:705–709
Kasemsumran S, Keeratinijakal V, Thanapase W, Ozaki Y (2010) Near infrared quantitative analysis of total curcuminoids in rhizomes of Curcuma longa by moving window partial least squares regression. J Near Infrared Spectrosc 18:263–269
Kasemsumran S, Apiwatanapiwat W, Suthiwijitpukdee N, Vaithanomsat V, Thanapase W (2014) Evaluation of Fourier transform-near infrared spectroscopic measurements for the quantification of curcumin in turmeric herbal medicines. J near Infrared Spectrosc 22:113–120
Kasemsumran S, Suthiwijitpukdee N, Keeratinijakal V (2017) Rapid classification of turmeric based on DNA fingerprint by near-infrared spectroscopy combined with moving window partial least squares-discrimination analysis. Anal Sci 33:111–115
Kim YJ, Lee HJ, Shin HS, Shin Y (2014) Near-infrared reflectance spectroscopy as a rapid and non-destructive analysis tool for curcuminoids in turmeric. Phytochem Anal 25:445–452
Lai X, Li J, Gong X, Lin X, Tang G, Li R, Jia C, Wang D, Ji S (2018) Rapid simultaneous determination of andrographolides in Andrographis paniculata by near-infrared spectroscopy. Anal Lett 51(17):2747–2762
Lee MS, Hwang YS, Lee J, Choung MG (2014) The characterization of caffeine and nine individual catechins in the leaves of green tea (Camellia sinensis L.) by near-infrared reflectance spectroscopy. Food Chem 158:351–357
Lim J, Mo C, Noh SH, Kang S, Lee K, Kim MS (2012) Capsaicinoids content prediction model development for Korean red-pepper powder using a visible and near-infrared spectroscopy. Proceeding of SPIE 8369, Sensing for agriculture and food quality and safety IV, 836901. https://doi.org/10.1117/12.977869
Liu J, Chen N, Yang J, Ouyang Z, Wu C, Yuan Y, Wang W, Chen M (2018) An integrated approach combining HPLC, GC/MS, NIRS, and chemometrics for the geographical discrimination and commercial categorization of saffron. Food Chem 253:284–292
Luypaert J, Zhang MH, Massart DL (2003) Feasibility study for the use of near infrared spectroscopy in the qualitative and quantitative analysis of green tea Camellia sinensis (L). Anal Chim Acta 478(2):303–312
Maeda H, Ozaki Y, Tanaka M, Hayashi N, Kojima T (1995) Near infrared spectroscopy and chemometrics studies of temperature-dependent spectral variations of water: relationship between spectral changes and hydrogen bonds. J near Infrared Spectrosc 3(4):191–201
McGoverin CM, Weeranantanaphan J, Downey G, Manley M (2010) The application of near infrared spectroscopy to the measurement of bioactive compounds in food commodities. J near Infrared Spectrosc 18(2):87–111
Murugan NA, Pandian CJ, Jeyakanthan J (2020) Computational investigation on Andrographis paniculata phytochemicals to evaluate their potency against SARS-CoV-2 in comparison to known antiviral compounds in drug trials. J Biomol Struct Dyn 29:1–12
Pholphana N, Rangkadilok N, Thongnest S, Ruchirawat S, Ruchirawat M, Satayavivad J (2004) Determination and variation of three active diterpenoids in Andrographis paniculata (Burm.f.) Nees. Phytochem Anal 15(6):365–371
Puri A, Axena R, Saxena RP, Saxena KC, Srivastava V, Tandon JS (1993) Immunostimulant agents from Andrographis paniculata. J Nat Prod 56:995–999
Rajani M, Shrivastava N, Ravishankara MN (2000) A rapid method for isolation of andrographolide from Andrographis paniculata Nees (Kalmegh). Pharm Biol 38:204–209
Ren G, Chen F (1999) Simultaneous quantification of ginsenosides in American ginseng (Panax quinquefolium) root powder by visible/near-infrared reflectance spectroscopy. J Agric Food Chem 47(7):2771–2775
Sa-ngiamsuntorn K, Suksatu A, Pewkliang Y, Thongsri P, Kanjanasirirat P, Manopwisedjaroen S, Charoensutthivarakul S, Wongtrakoongate P, Pitiporn S, Khemawoot P, Chutipongtanate S, Borwornpinyo S, Thitithanyanont A, Hongeng S (2021) Anti-SARS-CoV-2 activity of Andrographis paniculata extract and its major component Andrographolide in human lung epithelial cells and cytotoxicity evaluation in major organ cell representatives. J Nat Prod 84(4):1261–1270
Schulz H, Engelhardt UH, Wegent A, Drews HH, Lapczynski S (1999) Application of near-infrared reflectance spectroscopy to the simultaneous prediction of alkaloids and phenolic substances in green tea leaves. J Agric Food Chem 47(12):5064–5067
Sharma M, Sharma R (2013) Identification, purification and quantification of andrographolide from Andrographis paniculata (Burm. f.) Nees by HPTLC at different stages of life cycle of crop. J Curr Chem Pharm Sci 3:23–32
Sharma M, Sharma A, Tyagi S (2012) Quantitative HPLC analysis of andrograholide in Andrographis paniculata at two different stages of life cycle of plant. Acta Chim Pharm Indica 2:1–7
Shen YC, Chen CF, Chiou WF (2000) Suppression of rat neutrophil reactive oxygen species production and adhesion by the diterpenoid lactone andrographolide. Planta Med 66:314–317
Shi TH, Huang YL, Chen CC, Pi WC, Hsu YL, Lo LC, Chen WY, Fu SL, Lin CH (2020) Andrographolide and its fluorescent derivative inhibit the main proteases of 2019-nCoV and SARS-CoV through covalent linkage. Biochem Biophys Res Commun 533(3):467–473
Tahir HE, Arslan M, Mahunu GK, Mariod AA, Wen Z, Xiaobo Z, Xiaowei H, Jiyong S, El-Seedi H (2020) Authentication of the geographical origin of Roselle (Hibiscus sabdariffa L) using various spectroscopies: NIR low-field NMR, and fluorescence. Food Control 114:107231
Tanaka K, Kuba Y, Sasaki T, Hiwatashi F, Komatsu K (2008) Quantitation of curcuminoids in curcuma rhizome by near-infrared spectroscopic analysis. J Agric Food Chem 56:8787–8792
Wang L, Lee FSC, Wang X (2007) Near-infrared spectroscopy for classification of licorice (Glycyrrhizia uralensis Fisch) and prediction of the glycyrrhizic acid (GA) content. LWT-Food Sci Technol 40:83–88
Workman J, JrL W (2007) Practice guide to interpretive Near Infrared Spectroscopy. CRC Press, New York
Xu T, Pan J, Zhao L (2008) Simultaneous determination of four andrographolides in Andrographis paniculata Nees by silver ion reversed-phase high-performance liquid chromatography. J Chromatogr Sci 46(8):747–750
Acknowledgements
This research was funded by Kasetsart University Research and Development Institute (KURDI), Bangkok, Thailand (Grant Number 133.57).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kasemsumran, S., Apiwatanapiwat, W., Ngowsuwan, K. et al. Rapid selection of Andrographis paniculata medicinal plant materials based on major bioactive using near-infrared spectroscopy. Chem. Pap. 75, 5633–5644 (2021). https://doi.org/10.1007/s11696-021-01746-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11696-021-01746-0