Abstract
We report preliminary evidence of a symbiotic parabasalian protist in the guts of Peruvian mimic poison frog (Ranitomeya imitator) tadpoles. This species has biparental care and egg-feeding of tadpoles, while the related R. variabilis consumes the ancestral detritus diet in their nursery pools. Each species’ diet was experimentally switched, in the field and lab. Analyses of gut gene expression revealed elevated expression of proteases in the R. imitator field egg-fed treatment. These digestive proteins came from parabasalians, a group of protists known to form symbiotic relationships with hosts that enhance digestion. Genes that code for these digestive proteins are not present in the R. imitator genome, and phylogenetic analyses indicate that these mRNA sequences are from parabasalians. Bar-coding analyses of the tadpole microbiomes further confirmed this discovery. Our findings indicate the presence of parabasalian symbiotes in the intestines of the R. imitator tadpoles, that may aid the tadpoles in protein/lipid digestion in the context of an egg diet. This may have enabled the exploitation of a key ecological niche, allowing R. imitator to expand into an area with ecologically similar species (e.g., R. variabilis and R. summersi). In turn, this may have enabled a Müllerian mimetic radiation, one of only a few examples of this phenomenon in vertebrates.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Symbiotic relationships between microorganisms and their hosts are common, and have played major roles in the ability of certain organisms to expand into new ecological niches (Moran et al., 2019). Microbial community compositional changes have been documented extensively for their roles in the digestive system, particularly in humans (Gilbert et al., 2015). The vertebrate digestive system is inhabited by a wide array of complex microbial communities that can differ greatly between species, populations and individuals (Kuziel & Rakoff-Nahoum, 2022; Youngblut et al., 2019). We note that the microbiome is not restricted to bacteria, but can also include eukaryotic microorganisms, such as protists (Čepička et al., 2017). These communities have been shown to influence the immune system, cooperate in food breakdown, and induce specific gene expression in intestinal cells (Bosch & McFall-Ngai, 2011; Cash et al., 2006; Hooper et al., 2002). Their various roles result from strong exposure to variation in the gut environment (Yang et al., 2021).
External factors, such as those found in an organism’s habitat, also strongly influence composition (Bletz et al., 2016). Selection may influence diet based on the functional ability of gut microbes to degrade specific molecules in the food source introduced to the gut (Brune & Dietrich, 2015; Kohl et al., 2014). In one study, animals [desert woodrats (Neotoma lepida)] feeding on toxic tannin rich plants contained specific tannin degrading bacteria in the gut (Kohl et al., 2016). Thus, diet-associated microbes may be selected as gut colonists according to their ability to digest or detoxify commonly available food items (Alberdi et al., 2016).
Gut colonists can alter the physical and biochemical environment in the gut, potentially manifesting as changes in patterns of host gene expression. Gut microbes can also express genes themselves (e.g. digestive proteins) that affect host digestive capabilities and this can greatly influence host fitness, adaptive abilities and the potential to colonize new environments (Alberdi et al., 2016). For example, Sommer et al., (2016) showed that seasonal differences in the microbiota of active (as opposed to hibernating) brown bears affected the ability of the hosts to accumulate fat. In another study, Chevalier et al. (2015) demonstrated that cold-adapted microbiota altered gene expression patterns in the intestines of host mice. Therefore, studying both microbial composition and gene expression can provide unique insights into the interactions influencing biological adaptation.
Transitions in the ability to digest specific food types mediated by the microbiome have been key drivers of evolutionary innovations in multiple taxa (Moran et al., 2019). For example, gut microbiota make difficult to digest food, such as plants and algae, available as sources of energy to groups with specialized microbiota in their guts, such as termites (Brune & Dietrich, 2015) and ruminant mammals (Flint et al., 2008). These organisms accomplish this through gut modification to accommodate host-restricted bacterial communities with enzymatic processes necessary to access nutrients.
Studies of such transitions in amphibians are rare, but may be particularly fruitful, because amphibians have independently evolved a wide array of reproductive strategies and associated ecological niches and feeding strategies (Furness et al., 2022; Nunes-De-almeida et al., 2021; Summers et al., 2006). For example, neotropical frogs of the family Dendrobatidae have evolved a variety of reproductive strategies and parental care, including the evolution of trophic egg-feeding, a strategy where females deposit unfertilized trophic eggs for their tadpoles (Brown et al., 2008a, b; Carvajal-Castro et al., 2021). To date, little research has been done on the physiological or microbiological changes in tadpoles associated with this behavior.
Within the dendrobatids, the genus Ranitomeya provides unique opportunities for comparative studies, because it includes several closely related and sympatric species. Notably, Ranitomeya imitator and Ranitomeya variabilis possess dramatically different modes of parental care and associated tadpole feeding strategies, despite sharing similar habitats. Ranitomeya variabilis have the ancestral trait of breeding in large phytotelmata (pools of water within terrestrial plants) with mosquito larvae, and less protein-rich nutrient sources inside pools such as available detritus and algae which tadpoles consume (Brown et al., 2008a, b). Ranitomeya imitator have evolved the ability to breed in tiny phytotelmata, and regularly feed their tadpoles protein and lipid rich unfertilized eggs, as other nutritional sources are lacking in such small pools (Brown et al., 2008a, b). The behavior of egg feeding is hypothesized to be more recently evolved, and the shift in feeding strategy likely allowed R. imitator to colonize its current range in spite of prior occupancy by other similar species (e.g. R. variabilis, R. fantastica and R. summersi) (Brown et al., 2008a, b; Symula et al., 2001; Yeager et al., 2012). With the evolution of egg feeding, R. imitator was able to utilize much smaller pools in different plants, without the resources required for tadpole growth and survival.
A key characteristic of this study system is that tadpoles of both species are able to survive on other foods, if available, although detritus in the small pools used by R. imitator is minimal and generally insufficient for tadpole growth and development (Brown et al., 2008b). Fundamental questions about the evolution of this novel behavior in R. imitator concern how it affected genetic, behavioral, microbial characteristics of tadpoles consuming this dramatically different diet, and vice-versa. There are several mechanisms that could have enabled or enhanced the ability to process this novel diet, including (1) enhanced facultative or constitutive increases in the expression of digestive enzymes in the guts of R. imitator tadpoles, or (2) an altered gut microbiome in R. imitator, with gut microbes that allow better digestion of eggs. We note that these are not mutually exclusive possibilities, and both could be acting in concert.
To investigate the potential mechanisms underlying the evolution of egg feeding in this genus of frogs we used each species’ facultative ability to utilize multiple sources of food and developed a comparative experiment (Fig. 1). We used a factorial design in which both R. imitator and R. variabilis were fed a detritus or egg diet. We then examined gene expression in the guts of these tadpoles, and conducted a thorough microbial screening using bar-coding. Our results suggest the presence of an unanticipated evolutionary mechanism.
Results
Gene Expression
Figure 1 shows the experimental design of the differential expression experiments analyzed for this study. Controlling for multiple comparisons, between egg-fed, natural pool and detritus-fed, large pool treatments of R. imitator, 15 transcripts were significantly differentially expressed (Table 1).
BLAST (Camacho et al., 2009) searches (blastn) of the NCBI nucleotide database using these sequences matched sequences of protein digesting enzymes (e.g. cysteine peptidases) from a group of protists known as parabasalians (Čepička et al., 2017) for seven out of the fifteen sequences. For example, one transcript is most closely related to a cysteine peptidase from the parabasalian protists Trichomonas vaginalis (a well-known parasite of the human reproductive tract). Placing this sequence in a phylogenetic context using SHOOT (Emms & Kelly, 2022): (S1 Appendix, Materials and Methods) showed (Fig. 2) that this sequence falls within a clade of Trichomonas vaginalis sequences (sequence data for this species is abundantly available in the GenBank database). Six other transcripts also matched proteases (peptidases) closely related to proteases previously identified in parabasalian protists (particularly Trichomonas vaginalis, Trichomonas gallinae, Tritrichomonas foetus, Dientamoeba fragilis).
Phylogenetic tree depicting the evolutionary relationships of a sample peptidase sequence from the R. imitator transcriptomic sequencing (RNA-seq) gene expression results to other organisms in the nucleotide sequence database (see text for further explanation). Numbers at the base of each node represent bootstrap values. Letters and numbers following each species name represent individual sample sequence accession numbers
These parabasalian peptidase sequences were highly expressed in the field egg-fed treatment, but not in the field detritus-fed treatment. For example, Fig. 3 shows the expression levels for a cathepsin-like cysteine peptidase under the field egg-fed versus field detritus fed R. imitator. This gene was highly expressed in the R. imitator egg-fed treatment, but showed no expression in the detritus-fed treatment. The protein-digesting function of these enzymes is of obvious significance given the high protein content of an egg-based diet.
We also found increased expression of common gene products (actin, ITS, 5.8S, 18S and 28S rRNA) in the egg-fed treatment that also are most similar to sequences of parabasalian protist sequences. One of these transcripts was an actin gene that matched most closely to an actin sequence from the parabasalian Trichomitus batrachorum. This is the only parabasalian originally identified in amphibians, although it has now been identified in other ectotherms (Dobell, 1909). Taken together, these results suggest a significant contribution to gut gene expression in the presence of an egg diet from an unknown parabasalian in R. imitator. We note here that the 18S gene is used for the eukaryotic microbiome barcoding analyses that we describe below.
Some “host” (i.e. amphibian) genes were also found to be differentially expressed between R. imitator treatments. These included EF-hand calcium binding domain 14, golgi associated PDZ and coiled-coil motif containing (GOPC), and DnaJ heat shock protein (Hsp40) member B6. The results of BLAST searches reveal that the differentially expressed R. imitator transcripts were closely related to other anurans (e.g. frogs from the genus Bufo or Rana), implying that they were likely produced by Ranitomeya imitator itself, rather than by a symbiont (Table 1).
As a control, we also set up five pools with R. imitator tadpoles in the lab, which were fed eggs, using the same protocols used for R. variabilis. Analyses of differential expression between R. imitator treatments in the field (either fed or not-fed) did not yield any genes showing significant differential expression.
Conversely, comparison of lab egg-fed and field detritus-fed treatments of R. variabilis tadpoles yielded a substantial number of differentially expressed genes. We note that it was not practical to have an egg-fed R. variabilis treatment in the field, and attempts to establish a detritus-fed treatment of R. variabilis in the lab failed due to mortality. Of the 2451 differentially expressed transcripts between the lab egg-fed and field detritus-fed treatments, a number of those up-regulated in the egg-fed treatment closely matched (based on our annotations) the sequences of genes associated with lipid processing. These include apolipoprotein A1, a major component of high-density lipoproteins intimately involved in cholesterol metabolism and well-known in the context of human cardiovascular disease. Another example is CYP51A1, a member of the cytochrome P450 group of enzymes. These enzymes are also heavily involved in the metabolism of cholesterol, steroids and other lipids. Up-regulation of these genes in the guts of the R. variabilis tadpoles fed on an egg-diet appears to be a response to the high lipid levels associated with that diet.
None of the differentially expressed genes were shared between R. variabilis and R. imitator. We did not see differential expression of any of the parabasalian genes seen in the R. imitator egg-fed versus detritus-fed comparison, and none of the parabasalian sequences found in R. imitator matched similar sequences in the R. variabilis transcriptome when they were used as queries (transcriptome searched with blastn after conversion to a BLAST database). These results suggest the proposed novel symbiotic relationship between R. imitator tadpoles and an unknown parabasalian likely does not extend to R. variabilis, and represents a novel, derived “trait” (association) in R. imitator.
Bacterial Microbiome Analyses
Indicator species analysis identified one Operational Taxonomic Unit (OTU) from R. variabilis treatments in the family Rikenellaceae (detritus-fed), and three in R. imitator treatments, from one unclassified bacteria (egg-fed), one Bacteroidaceae (detritus-fed), and one Desulfovibrionaceae (egg-fed). Analyses of diversity showed no significant differences. Bacterial species richness showed no difference in variability between the egg-fed and non-egg-fed treatments in R. imitator (p = 0.767) or R. variabilis (p = 0.100) (S1 Appendix, Table S1, Fig. S1). Bacterial diversity measured using Shannon Diversity Index (H) also showed no significant differences between treatments in either species (R. imitator: p = 0.167, R. variabilis: p = 0.079) (S1 Appendix, Table S1, Fig. S2). Simpson’s Evenness also showed no significant differences (R. imitator: p = 0.175 R. variabilis: p = 0.364) (S1 Appendix, Table S1, Fig. S3).
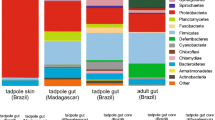
Plots of the results of the principal coordinate analyses of bacterial community composition for each species are shown in Figs. S4 and S5 (S1 Appendix). Results of the PERMANOVA analyses are summarized in Table S2 (S1 Appendix). Diet influenced bacterial communities of R. imitator (but not R. variabilis) (S1 Appendix, Table S2, Figs. S4, S5). Analyses of microbial community composition of R. variabilis and R. imitator uncovered bacteria aiding in digestion commonly found in the gut microbiome of many animals (Rikenellaceae in R. variabilis, Bacteroidaceae in R. imitator). The family found in R. variabilis was identified to genus level (Mucinivorans), which is known from one isolation from the digestive tract of a leech (Nelson et al., 2015), and could be associated with organisms digested by the tadpoles. Desulfovibrionaceae, found in egg-fed R. imitator, are composed of sulfate-reducing bacteria commonly found in aquatic environments often with high amounts of organic material. Some bacteria from this group have also been isolated from animal and human intestines, although their role in digestion is unknown.
Eukaryotic Microbiome Analyses
Similar to bacterial community patterns, microeukaryotic OTU richness, Shannon Diversity Indices, and Simpson’s Evenness Indices were similar across diets for each species (S1 Appendix, Figs. S6–S8). In addition, diet influenced microeukaryotic communities of R. imitator but not R. variabilis (S1 Appendix, Table S3, Figs. S9, S10).
We found two differentially expressed contigs in the R. imitator transcriptomic differential expression analyses that contained 18S sequences, which is the region used for our barcoding analyses of the eukaryotic microbiome. Using the blastn nucleotide-nucleotide search BLAST algorithm (Camacho et al., 2009), we searched the fasta file of the results from the microEuk 18S rRNA panel (as a BLAST database constructed in the program Geneious Prime 2022 (https://www.geneious.com) (S1 Appendix, Materials and Methods) using a query sequence from the 18S rRNA gene sequence that matched to Trichomonas gallinae. This search yielded one match, an 82 base pair sequence fragment from the R. imitator eukaryotic microbiome. We used this sequence in an unrestricted BLAST search (Camacho et al., 2009) of the main nucleotide database in GenBank. All the sequences returned (top 100 hits) were parabasalians. The top matches were with the parabasalians Tritrichomonas suis, T. foetus, T. nonconforma and T. augusta (100% query cover, Evalue 2e−26, percent match 95.12%). This result confirms the presence of parabasalians in the intestines of R. imitator tadpoles. It is likely that the exact species is (or are) as yet undescribed, given that there has been no previous research on the intestinal microbiota of Ranitomeya tadpoles.
Discussion
Ranitomeya imitator evolved a new, ecologically important mode of parental care—facultative egg feeding—as a putative mechanism of niche partitioning. We designed an experiment to investigate the potential mechanisms underlying the evolution of egg feeding in this species. We used examinations of tadpole gene expression in combination with microbial screens in tadpoles fed on egg and detritus only diets to better understand the evolution and maintenance of this ecological adaptation. We find compelling evidence for the presence of gut microbial parabasalians driving gut gene expression patterns in the presence of egg diets in Ranitomeya imitator tadpoles, suggesting the presence of symbiotic gut microbiota playing a key role in this adaptation.
Our transcriptomic analyses of the guts of R. imitator tadpoles that were fed a natural diet of eggs (a derived diet shared with its sister species R. vanzolinii), or detritus, algae and insect larvae (the ancestral diet for this genus) identified differences in expression of genes that we tentatively identified as being from a group of single-celled eukaryotes known as the Parabasalia. Parabasalians are anaerobic flagellated protists, most of which are symbionts found in the intestinal tracts of vertebrate and insect hosts (Čepička et al., 2017). They are perhaps most well known as gut mutualists of termites (Brune & Dietrich, 2015), which contribute to the digestion of wood as part of the termite gut microbiome. The most well-studied parabasalians are those found in humans, such as Trichomonas vaginalis (a urogenitotract parasite), or in domesticated animals, such as Tritrichomonas foetus (a venereal parasite of cattle, but a harmless commensal in pigs) (BonDurant & Honigberg, 1994). The relative dearth of sequences from other parabasalian lineages which are likely to be more related to parabasalians found in tropical amphibians likely explains why these well-characterized but presumably distantly related species provided the closest matches to the differentially expressed sequences in our BLAST and SHOOT searches. Parabasalians have been identified in amphibians [e.g. Trichomitus batrachorum (Dobell, 1909)], but there is comparatively little sequence data available for these species. In order to test our hypothesis that differential expression between egg-fed and non-egg-fed R. imitator is primarily driven by parabasalians in the gut, we conducted an analysis of the eukaryotic microbiota in R. imitator. A BLAST search of the results from our eukaryotic microbiome panel database using a query sequence from our differential expression analyses (a sequence that a general BLAST search had revealed closely matched an 18S rRNA sequence from the parabasalian Trichomonas gallinae), identified close matches between the query sequence and a sequence in the database. A BLAST search of the GenBank general nucleotide database using that sequence (from the eukaryotic microbiome) revealed close matches to multiple parabasalians with high degrees of similarity. Hence, the eukaryotic microbiome data provides independent support for the presence of parabasalians in the guts of R. imitator tadpoles.
As the intestinal microbiomes of Amazonian poison frogs were virtually unstudied until now, it is likely that the sequences we identified are from an as-yet-undescribed species of symbiotic parabasalian inhabiting the guts of R. imitator tadpoles. Given that these transcript sequences align to parabasalians but not the R. imitator genome (Stuckert et al., 2021), it is likely that they are from gut microfauna rather than the R. imitator gut transcriptome. Thus, it is likely that the differential expression observed results from either different population densities of the parabasalian microfauna, the upregulation of these genes in gut parabasalia, or a combination of the two. It is therefore likely that these microorganisms are responding to the gut microenvironment as affected by the differential dietary treatments. The presence of these microorganisms in the R. imitator gut and their potential role in the digestion of lipid and protein rich eggs, coupled with a lack of evidence for this microorganism in R. variabilis, suggests that this is an evolved symbiosis between R. imitator and its gut parabasalia.
While some of the parabasalian genes were common genes not associated with nutrient digestion (e.g. actin), most were protein digestion enzymes (e.g. cathepsins). These genes are known to be key mediators of protein catabolism. In fact, they are key enzymes involved in the processing of high protein substrates, such as blood meals in ticks and other blood feeding animals (Alim et al., 2009; Santiago et al., 2017). Cathepsins are also involved in lipid processing (Thibeaux et al., 2018). Hence, these enzymes play key roles in the digestion of proteins and lipids, which would be valuable in processing the concentrated proteins and lipids associated with an egg diet.
As noted above, while parabasalians are well-known to be involved in digestive mutualisms in the digestive tracts of termites (where they help to digest cellulose), they have not (to our knowledge) been shown to engage in protein-digesting symbioses in other taxa. However, digestive symbioses involving the secretion of proteases have been found in a variety of taxa. For example, the burying beetle (Nicrophorus vespilloides) appears to have a symbiotic relationship with protease-secreting yeasts, which help to digest the flesh of the dead mice upon which the beetle’s offspring feed (Brinkrolf et al., 2021; Vogel et al., 2017). Some flies that utilize cadavers and carcasses for reproduction (e.g. the blow fly, Chrysomya megacephala) have symbiotic relationships (Bhattacherjee et al., 2022) with bacteria that secrete protein-digesting enzymes (e.g. Chryseobacterium artocarpi). Aphids appear to have a symbiotic relationship with bacteria (Serratia symbiotica), which secretes proteases that assist with the digestion of plant proteins (Skaljac et al., 2019). There does not seem to be any compelling reason that parabasalians could not evolve mutualisms based on the secretion of proteases, as many other taxa have.
Alternatively, it is possible that the relationship between R. imitator tadpoles and their gut-dwelling parabasalians could be commensal or parasitic. Parabasalians are known to engage in both of these types of relationships with vertebrate hosts (see references above). In these cases, however, the parabasalians are not involved in the digestion of host foodstuffs, which seems likely to be the case in the R. imitator tadpoles as these genes are only expressed in the presence of trophic eggs. We hypothesize that the presence of these parabasalians increases the digestion of lipids and/or proteins from trophic eggs, providing additional nutrients to tadpoles from the same food source. Experiments in culture with trophic feeder eggs to test the efficacy of parabasalid egg digestion, and to test whether they also express the genes we identified as important here would be key corroborating evidence.
Ultimately, in order to determine whether the relationship between this novel parabasalian and its tadpole host is, in fact, mutualistic, an experimental approach will be required. Specifically, it will be necessary to raise tadpoles with and without parabasalians in their digestive tracts, and measure the effects of these treatments on growth and survival rates. This may require the use of parabasalid inoculations or knockdowns to create tractable treatment groups to test these key hypotheses related to mutualism.
Another important avenue to pursue in this regard will be to determine if the unknown parabasalid is transmitted between parent and offspring (vertical transmission) via trophic egg feeding. Vertical transmission has long been known to select for less virulent, more mutualistic symbionts (Alizon et al., 2009; Ewald, 1987). In the cockroach/subsocial wood-feeding cockroach (Cryptocercus)/termite lineage, horizontal transmission of parabasalid protists in gregarious cockroaches (via coprophagy) is associated with a parasitic lifestyle (Nalepa, 2020). The transition to eusociality in termites (from common ancestry with cockroaches) is thought to have originated via the initiation of vertical transmission of parabasalid (and oxymonad) protists, via proctodeal trophallaxis in the common ancestor of Cryptocercus (the subsocial sister lineage of termites) and the termites (Nalepa, 2020). Cryptocercus have a nuclear family structure focused on monogamous pairs feeding their own offspring via proctodeal trophallaxis, and the mutualistic protists are passed from parents to offspring through this form of feeding (Nalepa, 2020). In turn, this mutualistic relationship was likely to have been a key component of the transition to eusociality in the termites (Nalepa, 2015). As noted above, R. imitator is one of the only frogs to show a nuclear family structure based on monogamous pairs, and feeding of offspring (via trophic eggs) is a key characteristic of this species. It would be a remarkable instance of convergence if this same mechanism (vertical transmission of symbionts via parental feeding of offspring) were associated with advanced sociality in two very distantly related lineages.
There are several weaknesses of this study. For example, we were unable to carry out a fully crossed design in our experiments. It proved to be impractical to implement an egg-feeding regime in the field for the Ranitomeya variabilis (with the ancestral detritus diet), and our attempts to implement a non-egg fed treatment for this species in the lab failed (the tadpoles did not survive). Hence, our results concerning differential gene expression and microbiome composition for the non-egg-feeding species (R. variabilis) cannot be confidently ascribed to environment versus dietary treatment, and may well involve effects from both. However, this result is not pertinent to the main value of the experiments on the non-egg-feeding species, which was to assess whether the parabasalian genes that we found to be differentially-expressed in the egg-feeding species are also found in the non-egg feeding species (they were not). Hence, our experiments on R. variabilis (the non-egg-feeding species) serve to demonstrate that this symbiosis does not occur in a species closely related to the egg-feeding species (that has the ancestral detritus-based diet).
Another issue is that we did not find differential expression of the parabasalian genes in the lab-based egg feeding treatment with R. imitator (the trophic egg-feeding species), only in the field-based egg-feeding treatment (relative to the field-based non-egg-feeding treatment for this species). However, gene by environment interactions are commonly observed in previous studies, and it is quite possible that in order to get a viable “starter culture” of the parabasalian symbiotes, R. imitator tadpoles need to be in their natural pools in the field.
In summary, the evidence for differential expression of parabasalian peptidase genes in the guts of R. imitator tadpoles feeding on an egg diet (compared to the ancestral detritus diet) implicates these protists as gut symbionts in these tadpoles. The high expression of proteolytic enzymes associated with the breakdown of proteins and lipids in other taxa further suggests that these protists are symbionts that specifically aid in the digestion of large quantities of proteins and lipids associated with an egg-based diet.
We believe this is evidence of a new form of symbiosis that provided a novel mechanism for a “key innovation” in the life history of R. imitator: the evolution of egg-feeding. This trait likely allowed R. imitator to greatly expand its geographic range into those of the (ecologically similar) northern species that it is currently sympatric with (e.g. R. variabilis, R. fantastica, R. summersi) by allowing this species to use tiny pools that provided insufficient nutrients to these other (detritus feeding) species, resulting in the formation of a large Mullerian mimicry complex (Stuckert et al., 2014a, b; Symula et al., 2001; Twomey et al., 2013; Yeager et al., 2012). Our results lead to several key questions for future research, including whether these parabasalians have coevolved with and are dependent on their amphibian hosts.
Materials and Methods
Field and Lab Experiments: Peru
Fieldwork was conducted at four field sites around Tarapoto, San Martin, Peru from May through August 2017. Lab experiments were conducted in an outdoor enclosure in Tarapoto. Taking advantage of each species’ facultative ability to utilize multiple sources of food, we developed an experimental design for the field study designed to compare responses at the genomic and microbial levels in tadpoles of each species (R. imitator and R. variabilis) when developing on a diet of infertile eggs, and when developing on a diet of algae, detritus, and mosquito larvae (S1 Appendix, Materials and Methods). Briefly, we completed three treatments for R. imitator (field egg-fed, field detritus-fed, lab egg-fed), and two for R. variabilis (field detritus-fed, lab egg-fed) (see Fig. 1).
Gut Transcriptome Analysis
After each experiment, tadpoles were collected and RNA was extracted from half of the total samples in each treatment (S1 Appendix, Materials and Methods). We assembled a single transcriptome per species using data from each experimental treatment. Transcriptome assembly was done using the Oyster River Protocol v2.2.7, with the results from three assemblers merged with the program Orthofuser (MacManes, 2018). Error correction and trimming were done with RCorrector 1.01 (Song & Florea, 2015) and trimmomatic (Bolger et al., 2014). Transcriptome quality was assessed with BUSCO 3.0.1 (Simão et al., 2015) and TransRate 1.0.3 (Smith-Unna et al., 2016). Transcript counts were pseudo-quantified using Kallisto 0.43.0 (Bray et al., 2016) and tested for differential expression in R version 3.4.2 (R Core Team, 2020) using Sleuth version 0.29.0 (Pimentel et al., 2017). Differentially expressed transcripts were searched against known sequences using a nucleotide search (blastn) in BLAST (NCBI). To further confirm the evolutionary relationships of the sequences of some of the identified candidate genes in R. imitator, we used SHOOT (Emms & Kelly, 2022), a program that searches a large database of sequence-based phylogenetic trees and places a query sequence into a phylogenetic context. Details of these analyses are provided in the supplemental materials (S1 Appendix, Materials and Methods). Additionally, we used blastn in BLAST (Camacho et al., 2009) to search for the five putative parabasalian genes (asparaginyl endopeptidase-like cysteine peptidase, cysteine protease 8, cathepsin L-like cysteine peptidase, a second transcript of cathepsin L-like cysteine peptidase, and cathepsin L-like cysteine proteinase precursor), as well as several “host” (amphibian) genes in the Ranitomeya imitator genome assembly (Stuckert et al., 2021).
Gut Prokaryotic Microbiome Analysis
We characterized tadpole gut microbiome composition via amplicon sequencing of the 16S rRNA gene. We extracted genomic DNA from using standardized kits and primer sets (S1 Appendix, Materials and Methods), to amplify the V4 region of the 16S subunit of the ribosomal RNA gene in bacteria and archaea (Caporaso et al., 2012).
Sequence assembly and analyses were done using a standard mothur pipeline (v1.40.1) (Schloss et al., 2009). We ran all statistical analyses in the R Environment (R Core Team, 2020). Intraspecific comparisons were made between egg-fed and detritus-fed samples of both R. imitator and R. variabilis. To visualize patterns of microbial community composition among the two treatments and species, we used principal coordinate analysis of the bacterial community composition based on the Bray–Curtis dissimilarity coefficient (S1 Appendix). The Adonis function in the vegan package (Oksanen, 2015) was used to run permuted analysis of variance (PERMANOVA) to test for clustering significance. We rarefied sequences prior to calculating bacterial richness, evenness, and diversity metrics. We conducted indicator species analysis to identify taxa representative of each diet for each species using the labdsv package (Roberts, 2016).
Gut Eukaryotic Microbiome Analyses
For eukaryotic microbiome sequencing, we conducted 18S rRNA amplicon sequencing designed by the Earth Microbiome Project (Caporaso et al., 2012) (S1 Appendix, Materials and Methods). Sequences were assembled and analyzed using a standard mothur pipeline (v1.48.0) (Schloss et al., 2009). We assembled contigs from paired end reads, trimmed low quality bases, aligned sequences to the Silva Database (Quast et al., 2013; SSURef v132) (S1 Appendix, Materials and Methods).
Data availability
Reads for the gene expression analyses have been deposited in the NCBI Sequence Read Archive, under ID PRJNA854962 (sequences to be released to the public upon publication).
Code availability
All code and data used in the transcriptome analyses and microbiome studies are located in a public GitHub repository (DOI: https://doi.org/10.5281/zenodo.7807357) and NCBI SRA BioProject ID PRJNA855478 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA855478/).
References
Alberdi, A., Aizpurua, O., Bohmann, K., Zepeda-Mendoza, L. Z., & Gilbert, M. T. P. (2016). Vertebrate gut metagenomes confer rapid ecological adaptation? Trends in Ecology and Evolution, 31, 689–699. https://doi.org/10.1016/j.tree.2016.06.008
Alim, M. A., Tsuji, N., Miyoshi, T., Islam, M. K., Hatta, T., & Fujisaki, K. (2009). Legumains from the hard tick Haemaphysalis longicornis play modulatory roles in blood feeding and gut cellular remodeling and impact on embryogenesis. International Journal of Parasitology, 391, 97–107.
Alizon, S., Hurford, A., Mideo, N., & Van Baalen, M. (2009). Virulence evolution and the trade-off hypothesis: History, current state of affairs and the future. Journal of Evolutionary Biology, 22, 245–259. https://doi.org/10.1111/j.1420-9101.2008.01658
Bhattacherjee, R., De, S., Sharma, G., Ghosh, S., Mishra, S., Suman, D. S., & Banerjee, D. (2022). Prevalence of mouthpart sensilla and protease producing symbiotic gut bacteria in the forensic fly Chrysomya megacephala (Fabricius, 1794): Insight from foraging to digestion. Acta Tropica, 229, 106380. https://doi.org/10.1016/j.actatropica.2022.106380
Bletz, M. C., Goedbloed, D. J., Sanchez, E., Reinhardt, T., Tebbe, C. C., Bhuju, S., Geffers, R., Jarek, M., Vences, M., & Steinfartz, S. (2016). Amphibian gut microbiota shifts differentially in community structure but converges on habitat-specific predicted functions. Nature Communications, 7, 13699. https://doi.org/10.1038/ncomms13699
Bolger, A. M., Lohse, M., & Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics Oxford, England, 3015, 2114–2120. https://doi.org/10.1093/bioinformatics/btu170
BonDurant, R. H., & Honigberg, B. M. (1994). Trichomonads of veterinary importance. In J. P. Kreier (Ed.), Parasitic Protozoa (2nd ed., pp. 111–188). Academic Press.
Bosch, T. C., & McFall-Ngai, M. J. (2011). Metaorganisms as the new frontier. Zoology, 1144, 185–190. https://doi.org/10.1016/j.zool.2011.04.001
Bray, N., Pimentel, H., Melsted, P., & Pachter, L. (2016). Near-optimal probabilistic RNA-seq quantification. Nature Biotechnology, 34, 525–527. https://doi.org/10.1038/nbt.3519
Brinkrolf, K., Shukla, S. P., Griep, S., Rupp, O., Heise, P., Goesmann, A., Heckel, D. G., Vogel, H., & Vilcinskas, A. (2021). Genomic analysis of novel Yarrowia-like yeast symbionts associated with the carrion-feeding burying beetle Nicrophorus vespilloides. BMC Genomics, 22, 323.
Brown, J. L., Morales, V., & Summers, K. (2008a). Divergence in parental care, habitat selection and larval life history in two species of Peruvian poison frogs: An experimental analysis. Journal of Evolutionary Biology, 21, 1534–1543.
Brown, J. L., Twomey, E., Morales, V., & Summers, K. (2008b). Phytotelm size in relation to parental care and mating strategies in two species of Peruvian poison frogs. Behaviour, 1459, 1139–1165.
Brune, A., & Dietrich, C. (2015). The gut microbiota of termites: Digesting the diversity in the light of ecology and evolution. Annual Review of Microbiology, 69, 145–166. https://doi.org/10.1146/annurev-micro-092412-155715
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., & Madden, T. L. (2009). BLAST+: Architecture and applications. BMC Bioinformatics, 10, 421. https://doi.org/10.1186/1471-2105-10-421
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Huntley, J., Fierer, N., Owens, S. M., Betley, J., Fraser, L., Bauer, M., Gormley, N., Gilbert, J. A., Smith, G., & Knight, R. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME Journal, 68, 1621–1624. https://doi.org/10.1038/ismej.2012.8
Carvajal-Castro, J. D., Vargas-Salinas, F., Casas-Cardona, S., Rojas, B., & Santos, J. C. (2021). Aposematism facilitates the diversification of parental care strategies in poison frogs. Scientific Reports, 11, 19047. https://doi.org/10.1038/s41598-021-97206-6
Cash, H. L., Whitham, C. V., Behrendt, C. L., & Hooper, L. V. (2006). Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science, 3135790, 1126–1130.
Čepička, I., Dolan, M. F., & Gile, G. H. (2017). Parabasalia. In J. M. Archibald, A. G. B. Simpson, & C. H. Slamovits (Eds.), Handbook of the Protists (2nd ed., pp. 1175–1218). Springer.
Chevalier, C., Stojanović, O., Colin, D. J., Suarez-Zamorano, N., Tarallo, V., Veyrat-Durebex, C., Rigo, D., Fabbiano, S., Stevanović, A., Hagemann, S., Montet, X., Seimbille, Y., Zamboni, N., Hapfelmeier, S., & Trajkovski, M. (2015). Gut microbiota orchestrates energy homeostasis during cold. Cell, 163(6), 1360–1374.
Dobell, C. C. (1909). Researches on the intestinal protozoa of frogs and toads. Quarterly Journal of Microbial Science, 53, 201.
Emms, D. M., & Kelly, S. (2022). SHOOT: Phylogenetic gene search and ortholog inference. BMC Genome Biology, 23, 85.
Ewald, P. W. (1987). Transmission modes and evolution of the parasitism–mutualism continuum. Annals of the New York Academy of Sciences, 503, 295–306. https://doi.org/10.1111/j.1749-6632.1987.tb40616.x
Flint, H. J., Bayer, E. A., Rincon, M. T., Lamed, R., & White, B. A. (2008). Polysaccharide utilization by gut bacteria: Potential for new insights from genomic analysis. Nature Reviews Microbiology, 6(2), 121–131.
Furness, A. I., Venditti, C., & Capellini, I. (2022). Terrestrial reproduction and parental care drive rapid evolution in the trade-off between offspring size and number across amphibians. PLoS Biology, 20(1), e3001495. https://doi.org/10.1371/journal.pbio.3001495
Gilbert, S. F., Bosch, T. C. G., & Ledón-Rettig, C. (2015). Eco-Evo-Devo: Developmental symbiosis and developmental plasticity as evolutionary agents. Nature Reviews Genetics, 16, 611–622.
Hooper, L. V., Midtvedt, T., & Gordon, J. I. (2002). How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annual Review Nutrition, 221, 283–307. https://doi.org/10.1146/annurev.nutr.22.011602.092259
Kohl, K. D., Stengel, A., & Dearing, M. D. (2016). Inoculation of tannin-degrading bacteria into novel hosts increases performance on tannin-rich diets. Environmental Microbiology, 186, 1720–1729.
Kohl, K. D., Weiss, R. B., Cox, J., Dale, C., & Dearing, M. D. (2014). Gut microbes of mammalian herbivores facilitate intake of plant toxins. Ecology Letters, 1710, 1238–1246. https://doi.org/10.1111/ele.12329
Kuziel, G. A., & Rakoff-Nahoum, S. (2022). The gut microbiome. Current Biology, 32, R257–R264.
MacManes, M. D. (2018). The Oyster River Protocol: A multi-assembler and kmer approach for de novo transcriptome assembly. PeerJ, 6, e5428. https://doi.org/10.7717/peerj.5428
Moran, N. A., Ochman, H., & Hammer, T. J. (2019). Evolutionary and ecological consequences of gut microbial communities. Annual Review of Ecology, Evolution and Systematics, 50, 451–475.
Nalepa, C. A. (2015). Origin of termite eusociality: Trophallaxis integrates the social, nutritional, and microbial environments. Ecological Entomology, 40, 323–335. https://doi.org/10.1111/een.12197
Nalepa, C. A. (2020). Origin of mutualism between termites and flagellated gut protists: Transition from horizontal to vertical transmission. Frontiers in Ecology and Evolution, 8, 14. https://doi.org/10.3389/fevo.2020.00014
Nelson, M. C., Bomar, L., Maltz, M., & Graf, J. (2015). Mucinivorans hirudinis gen. nov., sp. nov., an anaerobic, mucin-degrading bacterium isolated from the digestive tract of the medicinal leech Hirudo verbana. International Journal of Systematics and Evolutionary Microbiology, 65(3), 990–995.
Nunes-De-almeida, C. H. L., Haddad, C. F. B., & Toledo, L. F. (2021). A revised classification of the amphibian reproductive modes. Salamandra, 57, 413–427.
Oksanen, J. (2015). Vegan: Ecological diversity 1, 1–12. https://doi.org/10.1029/2006JF000545
Pimentel, H., Bray, N. L., Puente, S., Melsted, P., & Pachter, L. (2017). Differential analysis of RNA-seq incorporating quantification uncertainty. Nature Methods, 147, 687–690. https://doi.org/10.1038/nmeth.4324
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., & Glöckner, F. O. (2013). The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Research, 41(Database issue), D590–D596. https://doi.org/10.1093/nar/gks1219
R Core Team. (2020). R: A language and environment for statistical computing (R Foundation for Statistical Computing). https://www.R-project.org/
Roberts, D. W. (2016). Ordination and multivariate analysis for ecology package “labdsv”. https://cran.r-project.org/web/packages/labdsv/labdsv.pdf.
Santiago, P. B., de Araújo, C. N., Motta, F. N., Praca, Y. R., Charneau, S., Dourado Bastos, I. M., & Santana, J. M. (2017). Proteases of haematophagous arthropod vectors are involved in blood- feeding, yolk formation and immunity—A review. Parasites and Vectors, 10, 79. https://doi.org/10.1186/s13071-017-2005-z
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., & Weber, C. F. (2009). Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied Environmental Microbiology, 7523, 537–7541.
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V., & Zdobnov, E. M. (2015). BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics, 3119, 3210–3212. https://doi.org/10.1093/bioinformatics/btv351
Skaljac, M., Vogel, H., Wielsch, N., Mihaljovic, S., & Vilcinskas, A. (2019). Transmission of a transmission of a protease-secreting bacterial symbiont among pea aphids via host plants. Frontiers in Physiology, 10, 438. https://doi.org/10.3389/fphys.2019.00438
Smith-Unna, R., Boursnell, C., Patro, R., Hibberd, J. M., & Kelly, S. (2016). TransRate: Reference-free quality assessment of de novo transcriptome assemblies. Genome Research, 268, 1134–1144. https://doi.org/10.1101/gr.196469.115
Sommer, F., Ståhlman, M., Ilkayeva, O., Arnemo, J. M., Kindberg, J., Josefsson, J., Newgard, C. B., Fröbert, O., & Bäckhed, F. (2016). The gut microbiota modulates energy metabolism in the hibernating brown bear Ursus arctos. Cell Reports, 147, 1655–1661.
Song, L., & Florea, L. (2015). Rcorrector: Efficient and accurate error correction for Illumina RNA-seq reads. Gigascience, 4, 48.
Stuckert, A. M. M., Chouteau, M., McClure, M., LaPolice, T. M., Linderoth, T., Nielsen, R., Summers, K., & MacManes, M. D. (2021). The genomics of mimicry: Gene expression throughout development provides insights into convergent and divergent phenotypes in a Müllerian mimicry system. Molecular Ecology, 3016, 4039–4061.
Stuckert, A. M. M., Saporito, R. A., Venegas, P. J., & Summers, K. (2014a). Alkaloid defenses of co-mimics in a putative Müllerian mimetic radiation. BMC Evolutionary Biology, 141, 1–8.
Stuckert, A. M. M., Venegas, P. J., & Summers, K. (2014b). Experimental evidence for predator learning and Müllerian mimicry in Peruvian poison frogs Ranitomeya, Dendrobatidae. Evolutionary Ecology, 283, 413–426.
Summers, K., McKeon, C. S., & Heying, H. (2006). The evolution of parental care and egg size: A comparative analysis in frogs. Proceedings of the Royal Society of London B, 273, 687–692.
Symula, R., Schulte, R., & Summers, K. (2001). Molecular phylogenetic evidence for a mimetic radiation in Peruvian poison frogs supports a Müllerian mimicry hypothesis. Proceedings of the Royal Society of London B, 268, 2415–2421.
Thibeaux, S., Siddiqi, S., Zhelyabovska, O., Moinuddin, F., Masternak, M. M., & Siddiqi, S. A. (2018). Cathepsin B regulates hepatic lipid metabolism by cleaving liver fatty acid–binding protein. Journal of Biology, 2936, 1910–1923.
Twomey, E., Yeager, J., Brown, J. L., Morales, V., Cummings, M., & Summers, K. (2013). Phenotypic and genetic divergence among poison frog populations in a mimetic radiation. PLoS One, 82, e55443.
Vogel, H., Shukla, S. P., Engl, T., Weiss, B., Fischer, R., Steiger, S., Heckel, D. G., Kaltenpoth, M., & Vilcinskas, A. (2017). The digestive and defensive basis of carcass utilization by the burying beetle and its microbiota. Nature Communications, 8, 15186. https://doi.org/10.1038/ncomms15186
Yang, J., Yu, Z., Wang, B., & Ndayisenga, F. (2021). Gut region induces gastrointestinal microbiota community shift in Ujimqin sheep Ovis aries: From a multi-domain perspective. Environmental Microbiology, 23, 7603–7616.
Yeager, J., Brown, J. L., Cummings, M., Morales, V., & Summers, K. (2012). Testing for selection on color pattern in a Mimetic Radiation. Current Zoology, 58, 668–676.
Youngblut, N. D., Reischer, G. H., Walters, W., Schuster, N., Walzer, C., Stalder, G., Ley, R. E., & Farnleitner, A. H. (2019). Host diet and evolutionary history explain different aspects of gut microbiome diversity among vertebrate clades. Nature Communications, 10, 2200. https://doi.org/10.1038/s41467-019-10191-3
Acknowledgements
K. Summers acknowledges funding from the National Geographic Society (Grant #WW-R015-17) and the National Science Foundation (DEB-1655336). M. Muscarella acknowledges funding from the National Science Foundation (OIA-1826801 Supplemental Award). We thank Katie Weigel for invaluable assistance with the fieldwork in Peru. This work followed protocols approved by the ECU Animal Care and Use Committee (IACUC AUP D376). Permits for fieldwork in Peru, and collection and export permits, were provided by the Peruvian Ministry of Wildlife (RDG No. 0278-2017-SERFOR/DGGSPFFS/CITES No. 17 PE001718).
Author information
Authors and Affiliations
Contributions
KDW and KS designed this project with consultation from AP. KDW carried out the fieldwork and labwork for this project. KDW, AMMS, MM, AP and KS all carried out data analyses for this project. KDW and KS wrote the manuscript, with contributions, assistance and review from AMMS, AP and MM. All authors contributed to the design and production of the figures.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Weinfurther, K.D., Stuckert, A.M.M., Muscarella, M.E. et al. Evidence for a Parabasalian Gut Symbiote in Egg-Feeding Poison Frog Tadpoles in Peru. Evol Biol 50, 239–248 (2023). https://doi.org/10.1007/s11692-023-09602-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11692-023-09602-7