Abstract
Hybridization is a widespread phenomenon in animals, and hybrid heterosis/breakdown could be key processes determining the evolutionary dynamics of hybrids. Indeed, hybrids are not consistently disadvantaged compared to the parental lineages, as was historically assumed. Multiple processes could lead to performance differences between parental lineages and their hybrids. Despite many studies evaluated the performance of hybrids, a quantitative synthesis is required to assess the general pattern. Here we used meta-analytic and meta-regression approaches to quantify the fitness differences between parental lineages and their hybrids, and to identify possible processes that could lead to these differences. Specifically, we tested biological and methodological parameters that could determine differences in performance between hybrids and parental lineages. Hybrid performance was extremely variable across studies, being often significantly higher or lower compared to the mean performance of their parents. Nevertheless, the averaged hybrid performance was similar to the fitness of parental lineages, with differences across studies related to how performance was assessed. Genetic divergence between parental lineages, and the approach used to identify hybrids were the parameters most strongly related to variation in hybrid performance. Performance was lower for hybrids between distantly related lineages. Furthermore, study settings and the use of imprecise approaches for hybrid identification (e.g. morphology-based) can bias assessments of performance. Studies performed on wild populations and using genetic approaches for hybrid identification detected more often a decreased hybrid performance, compared to laboratory studies. We highlight the importance of appropriate settings for a realistic understanding of the evolutionary impacts of hybridization.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Historically, animal hybridization was often considered of limited importance because uncommon or restricted to sympatric areas where distinct genetics lineages come into secondary contact (Duckworth & Semenov, 2017; Schwenk et al., 2008). Nevertheless, genomic evidences are increasingly showing that hybridization and introgression are widespread phenomena that can play a crucial role in speciation, extinction, and adaptive radiations (Sakai et al., 2001; Seehausen, 2004; Mallet, 2005; Capblancq et al., 2015; Bay & Ruegg, 2017Kagawa &Takimoto 2018). Hybridization is now considered to be pervasive in animals, with major consequences on evolutionary processes (Atsumi et al., 2021; Ficetola & Stöck, 2016; Thompson et al., 2021). It was often hypothesized that hybrids are generally disadvantaged compared to parental lineages (Barton & Hewitt, 1985). However, the growing evidence of a major role of hybridization for evolutionary outcomes suggests that hybrids are not uniformly disadvantaged compared to parents (Arnold & Hodges, 1995). In fact, hybridization may lead to either decreased, increased, or similar fitness compared to parental lineages (Atsumi et al., 2021; Lohr & Haag, 2015). For example, hybridization can lead to an increase in F1 fitness compared to the fitness of parents and F2, termed hybrid vigor or heterosis (Chan et al., 2018; Chen, 2013), while hybrid breakdown can occur when hybridization results in a decrease in fitness from F1 to F2 or backcross generation, because of genetic incompatibility or for limited performance of hybrids in the environment (Allendorf et al., 2001; Barreto et al., 2015). Overall, the performance of hybrids compared to their parents can show multiple patterns, with multiple studies showing heterogeneous outcomes (e.g. Barreto et al., 2015; Casas et al., 2012; Gélin et al., 2019; Walsh et al., 2016).
There are several processes that can potentially determine the differences observed in performance between hybrids and their parental lineages, including true biological effects, and processes related to the methods used in studies. Among the biological effects, (1) the genetic distance between parental lineages probably plays a key role in hybrid performance (Atsumi et al., 2021; Coughlan et al., 2021; Coyne & Orr, 1998; R. Stelkens & Seehausen, 2009). An increase in genetic distance could increase heterosis, but too large genetic distances determine genetic incompatibility and can cause hybrid breakdown (Dobzhansky, 1937; Matute et al., 2010). Thus, the hybrid performance is expected to be highest when the genetic distance between parents is neither too small nor too large (Wei & Zhang, 2018). However, this issue is still largely uncertain, and a recent meta-analysis suggest that genetic divergence between parental species increases the probability of hybrids to have smaller traits size than both parents (Atsumi et al., 2021). (2) Different generations of a single cross can show different performance (Rhode & Cruzan, 2005). For instance, it is possible that first generation hybrids are characterized by heterozygote advantage, while later generations could suffer of hybrid breakdown (Burton, 1990; Dobzhansky, 1970; Ellison et al., 2008; Šimková et al., 2021). Nevertheless, there are many factors that determine performance differences among the different generations of the same cross. (3) The hybridization between native and invasive species can be a major mechanism in accelerating the speed of biological invasions (Dlugosch et al., 2015; Grabenstein & Taylor, 2018; Huxel, 1999), thus, it is possible that in systems involving successful invaders, hybridization with native lineage could lead to offspring with better performance (Huxel, 1999).
In addition to the biological effects, the methods used in studies assessing hybrid performance can influence the results of analyses. (4) Even though laboratory and field studies should ideally lead to consistent results (Hillebrand & Gurevitch, 2014; Mathis et al., 2003), some studies revealed poor agreement between field and laboratory researches (e.g.Bezemer & Mills, 2003; Joron & Brakefield, 2003). This discordance could be caused by multiple processes, including differences of ecological context and to stressful condition in the laboratory (Ficetola & De Bernardi, 2005). (5) Hybrids are often identified through characteristic morphological traits, but molecular analysis can better detect hybrid and introgression avoiding classification errors (e.g. Vanhaecke et al., 2012). (6) Hybrid performance can be assessed on the basis of a variety of traits (e.g. breeding success, morphology, behavior), and the same hybrid can have poorer, better, or similar performance compared to parental parents, depending on the considered traits. For example, hybrid partridges can lay larger clutches than their parental lineages, but also suffer a higher predation rate (Casas et al., 2012). Broad-scale analyses, assessing performance variation across multiple systems are needed to evaluate how these processes can influence the observed variation of performance between hybrids and their parental lineages.
In this study, we used meta-analytic and meta-regression approaches (Arnqvist & Wooster, 1995; Nakagawa & Santos, 2012) to evaluate differences in performance between hybrids and their parental lineages in animals, and investigate some of the possible predictors of these patterns. In fact, there are many studies using experimental data on hybrid performance relative to specific cross between populations or species in animals, but literature syntheses are required to identify the general effects of these factors. The meta-analytic approach allows us to gather several independent studies to obtain general trends and conclusions on the animal hybrid performance. The aim of our study was to provide a quantitative synthesis on the hybrid performance compared to parental lineages, in order to identify how the different processes can determine variation across systems and studies. Specifically, we tested if differences in performance between hybrids and parental lineages are related to three potential biological processes: (1) genetic distance between parental lineages, (2) hybrid generations (i.e. F1 vs. backcrosses or other crosses), (3) effects of invasive species, and to three potential processes related to study design and approaches: (4) lab vs. field studies, (5) hybrid identification method (6) traits considered for analyses.
Methods
Literature Search and Selection Criteria
To obtain journal articles reporting hybrid performance, we performed a systematic literature research in Web of Science database using the key words “hybrid” and “fitness” with no restriction on publication year. Even though the term “fitness” refers to the breeding success of individuals, many ecological and evolutionary studies use this term when assessing differences for a very broad range of performance measures. Because these terms have a broad meaning and can be used in different contexts, we refined the research selecting Web of Science categories particularly relevant for evolutionary or ecological studies: ecology, evolutionary biology, genetics heredity, computer sciences interdisciplinary applications, multidisciplinary sciences, biology, zoology, entomology, marine freshwater biology, environmental sciences, fisheries, biodiversity conservation, forestry, behavioural sciences, ornithology, oceanography, water resources, physiology, reproductive biology, developmental biology. The literature research was performed on May 4th 2020 and produced 1595 journal articles which were screened in several steps (Fig. 1a). We examined each article to verify eligibility to the selection criteria for inclusion in the meta-analysis (Fig. 1b). The following criteria for data inclusion were adopted:
-
1.
Only studies focusing on animal hybrids were included
-
2.
We selected studies that report at least one quantitative comparison between one hybrid and one parental population, obtained with a statistical analysis that can be converted into an effect size. If no effect sizes were available, but raw data were obtainable, we extracted data directly from text, plots, or tables (average ± the amount of variation or dispersion, and sample size) and subsequently converted into an effect size. We used the ImageJ software to extract data from the plots (Schindelin et al., 2015).
-
3.
We only used comparisons of traits representing hybrid performance. Morphological and behavioural characters were considered when they could be interpreted in terms of performance (e.g. differences in body condition, growth rate, foraging ability).
-
4.
We exclude studies about parasitism, which were analysed in a dedicated review (Theodosopoulos et al., 2019).
The comparison between the performance of hybrids can be performed using different approaches, each of which has its own merit and limitations. In the mid-parent approach, the performance of hybrids is compared with the average value of parental lineages (Atsumi et al., 2021; Thompson et al., 2021). This approach is used to test the null-hypothesis that hybrids have intermediate performance compared to the parental species. The mid-parent approach maximizes the probability of detecting additive or non-additive genetic effects determining whether hybrid traits are intermediate, biased toward one parent (dominance) or a novelty compared to those of their parents. Conversely, other studies compared hybrid performance with the performance of parental species separately (either with the performance of each parent, either with only one parent) (hereafter: separate-parent approach; e.g.: Debes et al., 2013; Duckworth & Semenov, 2017; Good et al., 2000; Liss et al., 2016). This approach does not test explicitly whether hybrid performance mismatches or matches with the mid-value of parental lineages, but has larger power at detecting general patterns of variation in hybrid performance compared to parental one and the main drivers of these patterns (e.g. Kleindorfer et al., 2014; Walsh et al., 2018). Furthermore, the majority of studies retrieved by the literature analysis used the separate-parent approach (see results), thus considering this approach allowed to include a larger number of tests in the meta-analysis, increasing statistical power.
For each study, if possible we extracted the effect size of the difference in performance between the hybrid and the mid-point between parental lineages (mid-parent approach), and of the difference in performance between the hybrid and each parental lineage (separate-parent approach).
We also extracted information about six biological and methodological parameters that could determine differences in performance between hybrids and parental populations from the collected journal articles. (1) To analyse genetic distance between parental lineages, we used two partially overlapping approaches. First, we discriminated between intra-specific and inter-specific crosses. Furthermore, to estimate the genetic distance between parental lineages we also used TIMETREE, which calculates the divergence time for a pair of taxa (http://www.timetree.org) (Kumar et al., 2017). Unfortunately, TIMETREE information was only available for a limited subset of species, and was unavailable for intraspecific crosses. (2) As hybrid generations, we considered the generations belonging to: F1, first generation of backcross (BC), and hybrid above F1 and BC1 (e.g. F2, BC2, hereafter F > 1). In some studies, F > 1 included multiple generations that were pooled as a single type of hybrid by the authors. (3) We determined whether crosses occurred between natives or between one native and one invasive population. As for methodological parameters, we distinguished between: (4) field or laboratory study (the setting in which hybrid were measured), (5) genetic and morphological hybrid identification and (6) trait category considered for comparisons. Many different traits were used for the comparisons between hybrid and parental lineages in studies, and thus traits were pooled in larger traits category: fitness (e.g. clutch size, survival, development success), morphological (e.g., fluctuating asymmetry, wing length, fin height), and behavioural (e.g. total duration of suckling, foraging technique, arrival rank for reproductive season).
Extraction of Effect Size Measures
For each comparison between hybrid and parental populations, we calculated the effect size as the difference in performance between hybrid and parental population. As effect size we used Fisher’s z; the more the Fisher’s z value was greater or lesser than zero, the greater the extent of differences between hybrid and parental lineages. Comparisons where hybrids showed lower performance compared to the parental lineages were coded as negative Fisher’s z value, and vice-versa. All the analyses were repeated considering both the mid-parent and the separate-parent comparisons between hybrids and parental species.
The mid-parent value was calculated as the average performance of the two parental lineages; furthermore, we calculated their combined standard deviations. We transformed the obtained mean and standard deviation in Fisher’s z and its variance (z-var) and we extracted one effect size for each comparison between hybrid and mid-parent value. For separate-parent approach, we transformed the statistics values reported in studies (F, t, R2, χ2, means, and standard deviation of populations) in Fisher’s z and its variance (z-var) using the compute.es package in R (Del Re, 2013). When the statistic reported was Z-value, we directly calculated Fisher’s z and its variance as: Fisher’s z = Z/√(n-3) and z-var = 1/(n-3) (Hartung et al., 2008). For one study, we converted d-value to Pearson’s correlation coefficient r and then we extracted the Fisher’s z and its variance using the compute.es package. For studies that did not report test statistics, we calculated the effect size from P-values. In many cases, one hybrid group was compared to two parental lineages. In these cases, we extracted one effect size for each comparison. Different comparison between the same hybrid group and the two parental lineages were then identified by the same identity (hereafter: hybrid ID).
Finally, we recorded whether each comparison showed statistical differences between hybrids and parental lineages.
Statistical Analyses
For each comparison approach (mid-parent and separate-parent), we calculated the Rosenberg’s fail-safe number to evaluate file-drawer bias. Rosenberg’s fail-safe number establishes the studies that should be added to the meta-analysis to make the difference between observed and expected no longer significant and it estimates the strength of the results of sampling bias meta-analysis. We used Egger’s regression test and Begg’s rank test to evaluate the occurrence of publication bias in the dataset as procedure to implement the funnel plot (Begg & Mazumdar, 1994; Egger et al., 1997). Finally, we quantified heterogeneity using I2 (Nakagawa & Santos, 2012).
Factors Potentially Affecting the Significance of Comparisons
We used a χ2 test to assess if the studies detected significant differences between hybrids and the mid-parent values more frequent than expected by chance. Subsequently, we ran two generalized linear mixed-effects models (GLMM) to analyse the factors related to the frequency of significant comparisons. First, we evaluated if the sign of the comparisons was different between significant and non-significant comparisons. The positive sign represented a better hybrid performance compared to the average of parental groups, while the negative sign represents the opposite. We thus fitted a binomial GLMM to assess if significant positive results were more frequent than negative ones, by including taxonomic group (genus), study identity and hybrid ID as random factors. A second binominal GLMM assessed whether the frequency of significant effect sizes was related to: relationships between parents, hybrid generations, alien populations in parental cross, laboratory or field study, hybrid identification method, and trait category as fixed factors. Also in this case, we included taxonomic genus, identity of the study and hybrid ID as random factors. Binomial GLMMs were run using the lme4 package in R; we used a likelihood-ratio test to assess the significance of fixed factors.
Meta-analysis
We implemented meta-analysis and meta-regression approaches in a Bayesian framework using generalized linear mixed models (MCMCglmm package in R) (Hadfield, 2010; Nakagawa & Santos, 2012). We fitted different mixed models with different aims. All MCMC models were run with 60,000 iterations, discarding the first 10,000 iterations as a burn-in and with a thinning interval of 24. We used the mev argument in the MCMCglmm function to consider 1/z-variance as a weight for the records (Hadfield & Nakagawa, 2010).
Overall Meta-analysis: Model of the Mean
For each comparison approach, first, in order to analyse the mean performance value of hybrid relative to their parents, we ran a model of the mean considering the effect sizes of all different comparisons of collected studies. This analysis allowed us to assess whether the average fitness of hybrids was higher or lower relative to their parents. The effect sizes of the comparisons (Fisher’s z) were used as dependent variables, no fixed effect was included, and three random factors were added: taxonomic genus, identity of the study, and hybrid ID.
Average Performance for Different Categories
In order to discriminate factors that may determine differences in hybrid performance relative to their parents, we categorized comparisons by different author methods, hybrid features, and parental cross characteristics. The same categories were used for both mid-parent and separate-parent approaches. We performed several models of the mean to test the mean value of the effect sizes in different subsets of data. The following subsets were considered: (1) relationships between parents (intraspecific vs. interspecific crosses, and genetic distance between parental species), (2) hybrid generations (F1, F > 1, and backcrosses), (3) presence of native vs. invasive populations in parental cross, (4) laboratory or field study, (5) hybrid identification method (genetics vs. morphology), (6) trait category used for comparisons. In addition, we run a separate model for each taxonomic group (class) of parental lineages for which we obtained effect sizes from at least three different genera. For hybrid identification, laboratory crosses between morphologically identified parents are expected to be more accurate than the morphological of hybrids, even though without genetic data on parental lineages collected in the field also laboratory crosses could be imprecise, for example because of an unknown amount of introgression. Therefore, we re-run the analysis of hybrid identification method, by splitting morphological identification in two different categories: controlled crosses conducted in laboratory without genetic identification vs. morphological recognition of hybrids. All mixed-effects models included only the intercept and three random factors: taxonomic genus, identity of the study and hybrid ID.
Meta-regression for Divergence Between Parental Lineages
To visualize how the divergence between parental lineages can affect the hybrid performance, we ran two meta-regressions for the mid-parent approach, and four meta-regressions for the separate-parent approach. In all the models, hybrid performance (Fisher’s z) was the dependent variable. In the two models of mid-parent approach we used as predictors the relationships between parents (expressed as intra or inter-specific cross), and the genetic distance between parents obtained with TIMETREE, respectively. For the separate-parent approach, in two models we used relationships between parents (intra or inter-specific cross) as predictor. In the first model we considered all the effect sizes of the comparisons as dependent variable and in the second one we only considered the effect sizes of genetically identified hybrids as dependent variable. These models were also re-run after excluding four articles where hybrids were compared with only one parent; results were nearly identical to the analysis including all the studies (Tab. S1a). These two analyses were then repeated considering the genetic distance between parents obtained with TIMETREE as predictor; this analysis was limited to interspecific crosses for which divergence time was available on the basis of TIMETREE data. In the third model, we used all the effect sizes as dependent variable and in the fourth model we considered the effect sizes of only genetically identified hybrids as dependent variable. Due to small sample size, for the mid-parent approach it was not possible analysing separately the hybrids identified with genetic tools.
Meta-regression: Factors Potentially Affecting Hybrid Performance
To determine the factors related to the variation of hybrid performance compared to parents, we run two multivariable generalized linear mixed models one for each comparison approach. In these analyses, we used relationships between parents (intra or inter-specific) as an estimate of the divergence between parents, inasmuch there was available data for all the comparisons included. Contrary, TIMETREE database had a limited sample size for the divergence times. We used as dependent variable all the effect sizes of the comparisons and six parameters as fixed effects: relationships between parents, hybrid generations, invasive species cross, field vs lab studies, hybrid identification method, trait category used for comparisons; taxonomic genus, identity of the study and hybrid ID were added as random factors.
Moreover, we run a third model using separate-parent approach considering only the effect sizes obtained from studies that used genetic hybrid identification methods. We used five parameters as fixed effects: relationships between parents, hybrid generations, invasive species cross, lab vs field studies and trait category used for comparisons. Taxonomic genus, identity of the study and hybrid ID were added as random factors. We also we re-run the analysis excluding four articles in which hybrids were compared with only one parent, and obtained identical results (Tab. S1b).
Results
We retained 33 studies (Appendix S1) assessing hybrid performance with comparisons between hybrid and mid-parent value. These studies included 357 comparisons and 32 different animal species belonging to 9 taxonomic classes. For the separate-parent value approach, we retained 60 articles including 982 comparisons between hybrid and each parental lineage separately (Appendix S1). Studies focused on 94 different animal species belonging to 11 taxonomic classes. Overall, 66.7% of the collected studies compared hybrid to each parent separately, 12% compared hybrid with mid-parent value, 22% used both methods and 7% compared hybrid with only one parent.
Frequency of Significant Comparisons
Among the 357 comparisons between hybrids and the mean performance value of both parental populations, in 125 cases hybrid showed performance significantly different from parents (P < 0.05), and in 232 cases there were no significant differences (P > 0.05). The frequency of significant comparisons was much greater than expected under randomness (χ2 = 67.7, df = 1, p < < 0.001; number of significant comparisons expected under randomness: 17.85). The frequency of studies showing a positive significant effect was similar to the frequency of studies showing a negative significant effect (χ2 = 0.971, df = 1, p = 0.615). For the mid-parent approach, both Egger’s regression test (b = − 0.022, 95% CI = − 0.094/− 0.05) and Begg’s rank test (Kendall's τ coefficient = 0.0123, p = 0.0014) suggested some publication bias. Furthermore, we detected a strong heterogeneity of performance differences between hybrid and their mid-parent value across studies (total I2 = 96.91%). Nevertheless, the file drawer analysis suggested that 3622 unpublished, non-significant comparisons between parental and hybrids would be required to reduce the frequency of significant relationships to values similar to what is expected under randomness.
For the separate-parent approach, in 465 out of 982 comparisons hybrids showed performance significantly different from a parental species, while in 517 cases the authors did not detect significant differences. For the separate-parent approach, neither Egger’s regression test (b = − 0.037, 95% CI = − 0.108/-0.034) nor Begg’s rank test (Kendall's τ coefficient = − 0.027, p = 0.241) suggested publication bias. Also in this case, we found strong heterogeneity across studies (total I2 = 98.13%).
Factors Potentially Affecting the Significance of Comparisons
Within the 125 significant comparisons between hybrid and mid-parent performance, in 48 cases hybrids showed a lower performance, while in 77 comparisons hybrids showed better performance than parental lineages. Hybrids originating from intraspecific comparisons were more frequently different from the mid-parent value, compared to hybrids originating from interspecific comparisons (binomial generalized linear mixed model: χ2 = 3.86, df = 1, p = 0.049). The frequency of significant studies was similar between studies considering: different hybrid generations (χ2 = 3.283, df = 2, p = 0.194), alien populations in parental cross (mid-parent: χ2 = 2.019, df = 1, p = 0.156), hybrid identification method (mid-parent: χ2 = 0.886, df = 1, p = 0.347) and trait category (mid-parent: χ2 = 3.49, df = 2, p = 0.174). Finally, the mid-parent approach detected more often significant differences between hybrids and parental lineages in field studies, compared to laboratory studies (χ2 = 6.957, df = 1, p = 0.008).
Average Difference in Performance Between Hybrid and Parental Lineages
Using the mid-parent approach, the meta-analytical models calculating the average effect size across all the studies (model of the mean) suggested that the average performance of hybrids was slightly higher than the performance of the respective parental lineages, while the separate-parent approach suggested a slightly lower value. However, for both approaches the credible intervals overlapped zero, indicating that the average differences in performance were extremely limited (mid-parent approach: z = 0.027, 95% CI = − 0.05/0.113; separate-parent approach: mean z = − 0.052, 95% CI = − 0.136/0.031).
Meta-analysis for Subsets of Comparisons
Using the mid-parent approach, we did not detect significant differences between the considered subsets of data. The effect size of performance differences between hybrids and mid-parent performance overlapped zero for all the categories: intra-specific vs inter-specific parental lineage crosses, all the different hybrid generations, crosses involving only-native vs non-native parental lineages, all the systematic classes of parental lineages, field and laboratory studies, morphological vs. genetic hybrid identification methods, and the species traits measured for the comparisons (Tab S2).
When we re-ran the meta-analytic models for different subset of our data using separate-parent approach, we obtained, considering the relationships between parents, that the credible interval of the effect size of performance differences between hybrids and parental lineages overlapped zero for both intra-specific and for inter-specific crosses (Fig. 2a). When considering the hybrid generations, the credible interval of effect size overlapped zero for F1 crosses and for backcrosses, while was slightly more negative for crosses of subsequent generations (F > 1) (mean z = -0.125, 95% CI = − 0.209/− 0.042; Fig. 2b). Moreover, the credible interval of the effect size overlapped zero for: both crossing involving only native and crosses between non-native parental lineages (Fig. 2c), all the systematic classes of parental lineages and (Fig. 2d) field and laboratory studies (Fig. 2e). The mean effect size was significantly smaller than zero for hybrids identified through genetic approaches (mean z = − 0.116, 95% CI = − 0.219/− 0.015), while for hybrids identified through morphology the mean effect overlapped zero (Fig. 2f). The credible interval overlapped zero also for hybrids generated from controlled crosses conducted in laboratory without genetic identification of parental lineages which, in the previous analysis, were attributed to the “identified through morphology” group (Fig. S2). Finally, average effect size was not affected by methodological differences, as the credible interval of the effect size overlapped zero for studies considering fitness, morphological, and behavioural traits (Fig. 2g). The frequencies of the subset categories used in the collected studies and the means with 95% confidence interval of the subsets effect size are available in Online Resource (Tab. S3, Fig. S1).
Means of the effect sizes, as the difference in performance between hybrid and each parental population, in different subset of data. Density plots showing the means of the effect sizes for: a relationships between parents, b hybrid generations, c presence of an invasive population in parental cross, d parent’s class, e laboratory vs field studies, f hybrid identification method, g trait categories used for the comparisons
Do Divergence Between Parental Lineage Affects Performance? Meta-regression
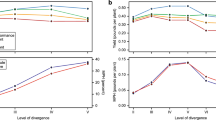
When considering all the effect sizes, there were no significant differences between intraspecific or interspecific hybrids using both comparison approaches (mid-parent approach: mean z = 0.03, 95% CI = − 0.144/ 0.205; separate-parent approach: mean z = − 0.054, 95% CI = − 0.247/ 0.121, Tab. S4). However, when we only considered hybrids identified through genetical approaches (409 comparisons), hybrids from interspecific crosses showed a lower performance than intraspecific hybrids (separate-parent approach: mean z = − 0.206, 95% CI = − 0.395/− 0.0195) (Table 1 a, b). Conversely, when we used the TIMETREE data to estimate interspecific divergence, we did not detect relationships between the amount of divergence and hybrid performance using both comparison approaches (mid-parent approach: mean z = − 0.026, 95% CI = − 0.103/− 0.044, Tab. S4; separate-parent approach: mean z = 0.002, 95% CI = − 0.009/− 0.014). Results were consistent considering effect size of hybrids only genetically identified (separate-parent approach: mean z = 0.002, 95% CI = − 0.014/0.018) (Tab. 1 c, d).
Overall Assessment of Factors Potentially Affecting Hybrid Performance
The meta-analysis including all the variables did not detect clear effects of any of the considered factors on hybrid performance comparing hybrid with both mid-parent value (Tab. S5) and each parent value separately (Table 2, Fig. 3a). Results were similar when we repeated the analysis only considering effect sizes obtained from studies that used genetic hybrid identification methods (Table 3, Fig. 3b).
Discussion
Despite long interest on hybrids, it remains difficult to identify a general trend for hybrid performance. By synthesizing 982 performance comparisons between hybrid and their parents, our meta-analysis provided insights on the role of several biological and methodological processes that could affect the outcome performance assessments. A large number of studies observed significant differences in performance between hybrids and parental lineages, and the variation in performance clearly was in different directions, with a comparable number of studies showing higher or lower performance, compared to parental lineages.
Two main approaches have been used to compare the fitness between hybrids and parental lineages, each of which can help identifying different facets of fitness variation during the hybridization process. Some studies have compared hybrid traits with the mid-parent value to investigate additive or nonadditive genetic effects of hybridization. However, this approach was only used by the minority of studies, focusing on the match or mismatch between hybrid performance and the intermediate features of parents (Atsumi et al., 2021; Thompson et al., 2021). The separate-parent approach was more common, because it can easily allow testing different patterns of hybrid performance, has less assumptions on the performance of hybrids, and does not require having accurate performance of both parental species. The two approaches yielded comparable conclusions, even though the mid-parent approach showed lower mean effect sizes compared to separate-parent approach (Fig. 4). Furthermore, the mid-parent approach allowed to include a lower number of studies than the separate-parent approach, and this reduced the statistical power of meta-analyses.
Generally, the average performance of hybrids was slightly lower than the one of their parents, but the differences in performance were extremely small, with strong heterogeneity across studies and approaches. Such heterogeneity is probably related to the very diverse processes that occur in different species, and can range from mortality and stillbirths, low viability, fertility, and survival (Fukui et al., 2018; Stelkens et al., 2015), to hybrid vigour, and adaptive advantages (Abbott et al., 2013; Meier et al., 2019). Such differences are probably linked to intrinsic differences across study systems, for instance to very different genetic architectures of animal species. Interactions between genotype and environment can also play an important role, thus the same system can show different outcomes depending on conditions experienced by individuals (Arnold & Martin, 2010; Grant & Grant, 1996). Furthermore, we found limited effect of the considered moderators on performance, and we only found some support for an effect of divergence between parental lineages, with hybrids between different lineages of the same species performing better than hybrid between different species.
Among the biological effects considered, we observed some support that the genetic distance between parents could be a driver of hybrid performance. Indeed, hybrids between distinct species showed lower performance than hybrids between lineages attributed to the same species (Table 1 a, b). This result might be related to the combined effects of heterosis and hybrid breakdown. Heterosis is often observed in hybrids between genetically close parents, and in some cases determine better fitness of hybrids (Atsumi et al., 2021; Dagilis et al., 2019). Our result aligns with a recent meta-analysis showing that genetically similar parents tend to produce hybrids with larger body size and reduced the phenotypic variability, while genetic distance between parental lineages increased this variability. Indeed, heterosis promotes developmental stability in hybrid between genetically close parental lineages (Atsumi et al., 2021). On the other hand, hybrids between different species often show hybrid breakdown. Hybrids between species can be inviable or sterile because the accumulation of genes that are regularly functional in pure-species, but produce negative epistatic interactions in hybrids. These postzygotic incompatibilities increase rapidly with the divergence between species (Dobzhansky, 1937). For instance, in Drosophila the amount of genes involved in hybrid breakdown increases with the divergence between two pairs of parental species (Matute et al., 2010). The genetic distance hypothesis would also predict that, for interspecific crossings, crossings involving distantly related species have lower fitness than the ones involving closely related species. However, we did not find evidence of relationships between relatedness (measured on the basis of TIMETREE) (Kumar et al., 2017) and hybrid performance (Table 1 c, d). This is partially in contrast with what we observed for the comparison intraspecific vs. interspecific crossings, and can be related to different causes. First, the TIMETREE data had a limited sample size, because genetic distances are not available for all the considered lineages. Furthermore, TIMETREE provides divergence time (in years) for a pair of taxa, but the time of divergence is not necessarily relevant as the same temporal divergence lead to a different genomic outcome depending, for example, on generation time and factors closely dependent on intrinsic characteristics of species (e.g. insects have much faster generations than vertebrates). Unfortunately, information on generation time is too scanty to be tested in this study.
It is known that different biological processes can affect hybrid performance and have been described quite well relative to a specific cross in the literature (e.g.: Campbell & Meinke, 2010; Casas et al., 2012). However, our meta-analysis did not identify a clear effect on these biological processes on hybrid performance, as only the divergence between parents affect hybrid performance. We found limited differences among hybrid generations, although it is known that F1 can be characterized by heterozygote advantage (Fitzpatrick & Bradley Shaffer, 2007). Nevertheless, advanced hybrid generations (F > 1) tended to have poorer fitness than e.g. F1 (Fig. 2b). These hybrids mostly represent a mix of different advanced generations (e.g. F2/F10), and under these conditions hybrids could suffer from hybrid breakdown, consequently this generation category showed lower performance than parents (Burton, 1990; Dobzhansky, 1970). In fact, the main hybrid breakdown is expected after F1 hybrid generation, when heterosis decreases and genetic incompatibilities increase (Dobzhansky, 1947). For instance, hybrid breakdown occurs in cichlid fish in F2 generation which shows particularly reduced fitness compare to parental species and F1 hybrids (Stelkens et al., 2015).
Finally, hybrids involving non-native lineages showed a performance similar to the ones only involving native lineages. Hybridization is often described as a major process determining the success of invasive alien species and it could lead to the loss of native populations through genetic pollution (Allendorf et al., 2001; Falaschi et al., 2020; Mooney & Cleland, 2001). For example, Italian Crested Newts, Triturus carnifex, were introduced in Western Switzerland, within the range of the native Great Crested Newts, T. cristatus. This introduction caused a massive introgression in Great Crested Newts and, sometimes, the total replacement of pure native species (Dufresnes et al., 2016). Hence, hybrids involving non-native lineages could have high performance in some ecological contexts (Ryan et al., 2009). Nevertheless, alien invasive species often show extremely high performance, for example at traits that allow to cope with novel environments or climate changes (Blackburn et al., 2009; Da Silva et al., 2021; Shik & Dussutour, 2020). Hence, in several cases the performance of native × introduced hybrid can be lower than the performance of the invasive parental species, even though higher or similar than the native parental line. Unfortunately, this expectation cannot be tested here, as most of native × introduced hybrid studies compared hybrid performance only with the native parental lineages. Finally, we did not detect differences in performance between native × introduced hybrid and native × native hybrid, inasmuch the strong data heterogeneity did not allow the delineation of a general trend.
When we analyzed the processes related to study design and methodology that could lead to different hybrid performance outcomes, we observed some difference between wild and laboratory hybrids. Compared to laboratory, in the wild hybrids showed a higher incidence of lower-performance comparisons. In the last decades, some debate existence on the consistence between the results obtained by field and laboratory studies. Laboratory studies have better control of experimental conditions (e.g. physiological and motivational variables), and limit the interactions with other species or individuals that can affect the outcomes of the experiment (Campbell et al., 2009). However, assays in captivity do not necessarily reflect the conditions in natural habitats, and laboratory environment could induce unpredictable effects and stressful conditions (Ficetola & De Bernardi, 2005; Joron & Brakefield, 2003; Niemelä & Dingemanse, 2014). Conversely, field studies avoid the removal of animals from their natural context and artificial responses of individuals to unnatural stimulations (Fisher et al., 2015; Osborn & Briffa, 2017). Nevertheless, field studies can be affected by uncontrollable environmental variation (Campbell et al., 2009), and can have limited replication levels because they are sometimes expensive in terms of money and time, or because of the complexity to tagging, tacking, and monitoring wild animals (D. L. M. Campbell et al., 2009; Fisher et al., 2015). Furthermore, measuring individual performance in the wild is challenging and, without genetic data, it is difficult to ascertain the introgression status of individuals. Some analyses revealed poor agreement between field and laboratory researches (e.g.Bezemer & Mills, 2003; Joron & Brakefield, 2003), while others suggested that laboratory studies provide a good representation of patterns occurring in the wild (Hillebrand & Gurevitch, 2014; Mathis et al., 2003). However, we did not detect clear differences between these study typologies, supporting the idea that well-planned laboratory studies can provide results consistent with what is observed in the wild (e.g.: Herborn et al., 2010). In the context of hybrid performance studies, both lab and field studies have their own advantages, and the selection of the most appropriate approach can be dictated by species-specific technical constraints (e.g. feasibility of studying animals in the lab vs. in the lab), as well as by study aims.
The same hybrid can have lower, upper or similar performance compared to parental lineages based on the trait category considered for comparisons (e.g. breeding success, morphology, behavior…) (e.g. Campbell & Meinke, 2010; Casas et al., 2012; Gélin et al., 2019). For instance, Bryden et al., (2004) examined 12 performance traits in Chinook salmon comparing hybrid and parental lineages. Introgressed salmons showed better performance at growth-related traits, but also a poor resistance to pathogens. Although such differences can be easily determined for specific crosses, the meta-analytic approach failed to find for which trait category hybrids are more or less performing compared to parental lineages, probably because of the huge variety of investigated traits across studies or species.
We found differences between studies using genetic vs. non-genetic approaches for the identification of parental lineages and their hybrids. Hybrids and parental lineages showed similar performance if only morphology was used to identify hybrids, while the lower performance of hybrids was evident in studies using genetic identification. In several cases, it is extremely difficult to identify pure or hybrid lineages in absence of genetic data, thus these differences can be related to misidentification that can occur using morphological approaches (Dowling et al., 2015). Phenotypic traits are not always a reliable diagnostic method to recognize different lineages as they can vary strongly depending on the life histories (Vanhaecke et al., 2012). For instance, widely proposed morphological approach do not allow the perfect discrimination between the marsh frog (Pelophylax ridibundus) and the hybrid edible frogs (Pelophylax kl. esculentus), inasmuch several morphological characters greatly overlap between them (Pagano & Joly, 1999). Our results highlight the importance of genetic analyses for the correct identification of hybrids and avoid classification errors. Only after we limited our analyses to genetically-identified hybrids, we detected a negative relationship between genetic distance of parental lineages and the performance of hybrids (Table 1b). Thus, hybrid identification through genetic methods provides higher power to any kind of analysis. The growing availability and decreasing cost of genetic markers now enables fast identification of hybrids even in complex situation (Della Croce et al., 2016). Genetic analysis can also detect different rates of introgression in individuals, even when low introgression occurs. In fact, the amount of introgression can elicit different performance of the hybrids, and the extent or the direction of introgression can lead to different hybridization outcomes (Aboim et al., 2010; Payseur, 2010). For instance, in some systems the performance of hybrids can decrease at increasing proportion of introgression (Muhlfeld et al., 2009).
In conclusion, hybrid performance can be extremely variable. Hybrids often show significantly different performance compared to their parental lineages, still the very strong heterogeneity across studies makes it difficult to determine a general pattern of performance variation. Here, we have shown how both biological (genetic divergence) and methodological (hybrid identification method) factors may influence the detected hybrid performance. Hence, heterosis and hybrid breakdown could play a key role in the evolutionary dynamics of animal hybrids, and genetic approaches are fundamental to improve our understanding of these complex systems. Despite the huge amount of work on hybrid systems in the last decades, we are far from exhaustive knowledge of the factors determining the variation of hybrid performance. Nevertheless, the growing methodological (e.g. genomic analyses) and conceptual developments are opening new study avenues that can improve our understanding of hybridization as major component of the evolutionary processes.
Data Availability
Data generated or analysed during this study are included in this article and its supplementary information files.
References
Abbott, R., Albach, D., Ansell, S., Arntzen, J. W., Baird, S. J. E., Bierne, N., et al. (2013). Hybridization and speciation. Journal of Evolutionary Biology, 26(2), 229–246. https://doi.org/10.1111/j.1420-9101.2012.02599.x
Aboim, M. A., Mavárez, J., Bernatchez, L., & Coelho, M. M. (2010). Introgressive hybridization between two Iberian endemic cyprinid fish: A comparison between two independent hybrid zones. Journal of Evolutionary Biology, 23(4), 817–828. https://doi.org/10.1111/j.1420-9101.2010.01953.x
Allendorf, F. W., Leary, R. F., Spruell, P., & Wenburg, J. K. (2001). The problems with hybrids: Setting conservation guidelines. Journal of Archaeological Science, 6(11), 613–622. https://doi.org/10.1016/0305-4403(79)90038-4
Arnold, M. L., & Hodges, S. A. (1995). Are natural hybrids fit or unfit relative to their parents? Trends in Ecology & Evolution, 10(2), 67–71.
Arnold, M. L., & Martin, N. H. (2010). Hybrid fitness across time and habitats. Trends in Ecology and Evolution, 25(9), 530–536. https://doi.org/10.1016/j.tree.2010.06.005
Arnqvist, G., & Wooster, D. (1995). Meta-analysis: Synthesizing research findings in ecology and evolution. Trends in Ecology & Evolution, 10(6), 236–240. https://doi.org/10.1016/S0169-5347(00)89073-4
Atsumi, K., Lagisz, M., & Nakagawa, S. (2021). Nonadditive genetic effects induce novel phenotypic distributions in male mating traits of F1 hybrids. Evolution, 75(6), 1304–1315. https://doi.org/10.1111/evo.14224
Barreto, F. S., Pereira, R. J., & Burton, R. S. (2015). Hybrid dysfunction and physiological compensation in gene expression. Molecular Biology and Evolution, 32(3), 613–622. https://doi.org/10.1093/molbev/msu321
Barton, N. H., & Hewitt, G. M. (1985). Anlysis of hybrid zone. Annual Review of Ecology and Systematics, 16, 113–148.
Bay, R. A., & Ruegg, K. (2017). Genomic islands of divergence or opportunities for introgression? Proceedings of the Royal Society B: Biological Sciences, 284(1850), 20162414. https://doi.org/10.1098/rspb.2016.2414
Begg, C. B., & Mazumdar, M. (1994). Operating characteristics of a rank correlation test for publication bias. Biometrics, 50(4), 1088–1101.
Bezemer, T. M., & Mills, N. J. (2003). Clutch size decisions of a gregarious parasitoid under laboratory and field conditions. Animal Behaviour, 66(6), 1119–1128. https://doi.org/10.1006/anbe.2003.2296
Blackburn, T. M., Cassey, P., & Lockwood, J. L. (2009). The role of species traits in the establishment success of exotic birds. Global Change Biology, 15(12), 2852–2860. https://doi.org/10.1111/j.1365-2486.2008.01841.x
Bryden, C. A., Heath, J. W., & Heath, D. D. (2004). Performance and heterosis in farmed and wild Chinook salmon (Oncorhynchus tshawytscha) hybrid and purebred crosses. Aquaculture, 235(1–4), 249–261. https://doi.org/10.1016/j.aquaculture.2004.01.027
Burton, R. S. (1990). Hybrid breakdown in developmental time in the copepod Tigriopus californicus. Evolution, 44(7), 1814–1822. https://doi.org/10.1111/j.1558-5646.1990.tb05252.x
Campbell, D. L. M., Weiner, S. A., Starks, P. T., & Hauber, M. E. (2009). Context and control: Behavioural ecology experiments in the laboratory. Annales Zoologici Fennici, 46(2), 112–123. https://doi.org/10.5735/086.046.0204
Campbell, L. A., & Meinke, L. J. (2010). Fitness of Diabrotica barberi, Diabrotica longicornis, and their hybrids (Coleoptera: Chrysomelidae). Annals of the Entomological Society of America, 103(6), 925–935. https://doi.org/10.1603/AN10031
Capblancq, T., Després, L., Rioux, D., & Mavárez, J. (2015). Hybridization promotes speciation in Coenonympha butterflies. Molecular Ecology, 24, 6209–6222. https://doi.org/10.1111/mec.13479
Casas, F., Mougeot, F., Sánchez-Barbudo, I., Dávila, J. A., & Viñuela, J. (2012). Fitness consequences of anthropogenic hybridization in wild red-legged partridge (Alectoris rufa, Phasianidae) populations. Biological Invasions, 14(2), 295–305. https://doi.org/10.1007/s10530-011-0062-3
Chan, W. Y., Peplow, L. M., Menéndez, P., Hoffmann, A. A., & van Oppen, M. J. H. (2018). Interspecific hybridization may provide novel opportunities for coral reef restoration. Frontiers in Marine Science, 5, 1–15. https://doi.org/10.3389/fmars.2018.00160
Chen, Z. J. (2013). Genomic and epigenetic insights into the molecular bases of heterosis. Nature Reviews Genetics, 14(7), 471–482. https://doi.org/10.1038/nrg3503
Coughlan, J. M., Dagilis, A. J., Serrato-Capuchina, A., Elias, H., Peede, D., Isbell, K., et al. (2021). Population structure and introgression among recently differentiated. Biorxiv. https://doi.org/10.1101/2021.06.25.449842
Coyne, J. A., & Orr, H. A. (1998). The evolutionary genetics of speciation. Philosophical Transactions of the Royal Society B: Biological Sciences, 353(1366), 287–305. https://doi.org/10.1098/rstb.1998.0210
Da Silva, C. R. B., Beaman, J. E., Dorey, J. B., Barker, S. J., Congedi, N. C., Elmer, M. C., et al. (2021). Climate change and invasive species: a physiological performance comparison of invasive and endemic bees in Fiji. Journal of Experimental Biology, 224(1), jeb230326. https://doi.org/10.1242/jeb.230326
Dagilis, A. J., Kirkpatrick, M., & Bolnick, D. I. (2019). The evolution of hybrid fitness during speciation. PLoS Genetics, 15(5), 1–21. https://doi.org/10.1371/journal.pgen.1008125
Debes, P. V., Fraser, D. J., McBride, M. C., & Hutchings, J. A. (2013). Multigenerational hybridisation and its consequences for maternal effects in Atlantic salmon. Heredity, 111(3), 238–247. https://doi.org/10.1038/hdy.2013.43
Del Re, A. C. (2013). Compute.es: Compute effect sizes. R package version 0.2–2.
Della Croce, P., Poole, G. C., & Luikart, G. (2016). Detecting and quantifying introgression in hybridized populations: Simplifying assumptions yield overconfidence and uncertainty. Molecular Ecology Resources, 16(6), 1287–1302. https://doi.org/10.1111/1755-0998.12520
Dlugosch, K. M., Anderson, S. R., Braasch, J., Cang, F. A., & Gillette, H. D. (2015). The devil is in the details: Genetic variation in introduced populations and its contributions to invasion. Molecular Ecology, 24(9), 2095–2111. https://doi.org/10.1111/mec.13183
Dobzhansky, T. (1937). Genetics and the origin of species. Columbria University Press.
Dobzhansky, T. (1947). Genetic structure of natural populations. Washington: Carnegie Institute of Washington Year Book.
Dobzhansky, T. (1970). Genetics of the evolutionary process. Columbia University Press.
Dowling, T. E., Smith, G. R., & Brown, W. M. (2015). Reproductive isolation and Introgression between Notropis cornutus and Notropis chrysocephalus (Family Cyprinidae): comparison of morphology, Allozymes, and mitochondrial DNA. Evolution, 43(3), 620–634.
Duckworth, R. A., & Semenov, G. A. (2017). Hybridization associated with cycles of ecological succession in a passerine bird. American Naturalist, 190(4), E94–E105. https://doi.org/10.1086/693160
Dufresnes, C., Pellet, J., Bettinelli-Riccardi, S., Thiébaud, J., Perrin, N., & Fumagalli, L. (2016). Massive genetic introgression in threatened northern crested newts (Triturus cristatus) by an invasive congener (T. carnifex) in Western Switzerland. Conservation Genetics, 17(4), 839–846. https://doi.org/10.1007/s10592-016-0825-6
Egger, M., Smith, G. D., Schneider, M., & Mider, C. (1997). Bias in meta-analysis detected by a simple, graphical test. BMJ, 315(7109), 629–634.
Ellison, C. K., Niehuis, O., & Gadau, J. (2008). Hybrid breakdown and mitochondrial dysfunction in hybrids of Nasonia parasitoid wasps. Journal of Evolutionary Biology, 21(6), 1844–1851. https://doi.org/10.1111/j.1420-9101.2008.01608.x
Falaschi, M., Melotto, A., Manenti, R., & Ficetola, G. F. (2020). Invasive species and amphibian conservation. Herpetologica, 76(2), 216–227. https://doi.org/10.1655/0018-0831-76.2.216
Ficetola, G. F., & De Bernardi, F. (2005). Interspecific social interactions and breeding success of the frog Rana latastei: A field study. Ethology, 111(8), 764–774. https://doi.org/10.1111/j.1439-0310.2005.01089.x
Ficetola, G. F., & Stöck, M. (2016). Do hybrid-origin polyploid amphibians occupy transgressive or intermediate ecological niches compared to their diploid ancestors? Journal of Biogeography, 43(4), 703–715. https://doi.org/10.1111/jbi.12667
Fisher, D. N., James, A., Rodríguez-Muñoz, R., & Tregenza, T. (2015). Behaviour in captivity predicts some aspects of natural behaviour, but not others, in a wild cricket population. Proceedings of the Royal Society B: Biological Sciences, 282(1809), 20150708. https://doi.org/10.1098/rspb.2015.0708
Fitzpatrick, B. M., & Bradley Shaffer, H. (2007). Hybrid vigor between native and introduced salamanders raises new challenges for conservation. Proceedings of the National Academy of Sciences of the United States of America, 104(40), 15793–15798. https://doi.org/10.1073/pnas.0704791104
Fukui, S., May-McNally, S. L., Taylor, E. B., & Koizumi, I. (2018). Maladaptive secondary sexual characteristics reduce the reproductive success of hybrids between native and non-native salmonids. Ecology and Evolution, 8(23), 12173–12182. https://doi.org/10.1002/ece3.4676
Gélin, U., Keller, M., de Beaupuis, V., Nowak, R., Lévy, F., & Locatelli, Y. (2019). Impact of hybridization between sika and red deer on phenotypic traits of the newborn and mother–young relationships. Animal Behaviour, 158, 65–75. https://doi.org/10.1016/j.anbehav.2019.09.016
Good, T. P., Ellis, J. C., Annett, C. A., & Pierotti, R. (2000). Bounded hybrid superiority in an avian hybrid zone: Effects of mate, diet, and habitat choice. Evolution, 54(5), 1774–1783. https://doi.org/10.1111/j.0014-3820.2000.tb00721.x
Grabenstein, K. C., & Taylor, S. A. (2018). Breaking barriers: Causes, consequences, and experimental utility of human-mediated hybridization. Trends in Ecology and Evolution, 33(3), 198–212. https://doi.org/10.1016/j.tree.2017.12.008
Grant, B. R., & Grant, P. R. (1996). High survival of Darwin’ s Finch Hybrids: Effects of beak morphology and diets. Ecology, 77(2), 500–509.
Hadfield, J. (2010). MCMC methods for multi-response generalized linear mixed models: The MCMCglmm R Package. Journal of Statistical Software, 33(2), 1–22.
Hadfield, J. D., & Nakagawa, S. (2010). General quantitative genetic methods for comparative biology: Phylogenies, taxonomies and multi-trait models for continuous and categorical characters. Journal of Evolutionary Biology, 23(3), 494–508. https://doi.org/10.1111/j.1420-9101.2009.01915.x
Hartung, J., Knapp, G., & Sinha, B. K. (2008). Statistical meta-analysis with applications. N. J. John Wiley & Sons Inc.
Herborn, K. A., Macleod, R., Miles, W. T. S., Schofield, A. N. B., Alexander, L., & Arnold, K. E. (2010). Personality in captivity reflects personality in the wild. Animal Behaviour, 79(4), 835–843. https://doi.org/10.1016/j.anbehav.2009.12.026
Hillebrand, H., & Gurevitch, J. (2014). Meta-analysis results are unlikely to be biased by differences in variance and replication between ecological lab and field studies. Oikos, 123(7), 794–799. https://doi.org/10.1111/oik.01288
Huxel, G. R. (1999). Rapid displacement of native species by invasive species: Effects of hybridization. Biological Conservation, 89(2), 143–152. https://doi.org/10.1016/S0006-3207(98)00153-0
Joron, M., & Brakefield, P. M. (2003). Captivity masks inbreeding effects on male mating success in butterflies. Nature, 424(6945), 191–194. https://doi.org/10.1038/nature01713
Kagawa, K., & Takimoto, G. (2018). Hybridization can promote adaptive radiation by means of transgressive segregation. Ecology Letters, 21(2), 264–274. https://doi.org/10.1111/ele.12891
Kleindorfer, S., O’Connor, J. A., Dudaniec, R. Y., Myers, S. A., Robertson, J., & Sulloway, F. J. (2014). Species collapse via hybridization in Darwin’s tree finches. American Naturalist, 183(3), 325–341. https://doi.org/10.1086/674899
Kumar, S., Stecher, G., Suleski, M., & Hedges, B. (2017). TimeTree: A resource for timelines, timetrees, and divergence times. Molecular Biology and Evolution, 34(7), 1812–1819. https://doi.org/10.1142/9789814401241_0003
Liss, S. A., Lamer, J. T., Sass, G. G., & Suski, C. D. (2016). Physiological consequences of hybridization: Early generation backcrossing decreases performance in invasive bigheaded carps. Journal of Freshwater Ecology, 31(4), 543–554. https://doi.org/10.1080/02705060.2016.1188426
Lohr, J. N., & Haag, C. R. (2015). Genetic load, inbreeding depression, and hybrid vigor covary with population size: An empirical evaluation of theoretical predictions. Evolution, 69(12), 3109–3122. https://doi.org/10.1111/evo.12802
Mallet, J. (2005). Hybridization as an invasion of the genome. Trends in Ecology and Evolution, 20(5), 229–237. https://doi.org/10.1016/j.tree.2005.02.010
Mathis, A., Murray, K. L., & Hickman, C. R. (2003). Do experience and body size play a role in responses of larval ringed salamanders, Ambystoma annulatum, to predator kairomones? Laboratory and field assays. Ethology, 109(2), 159–170. https://doi.org/10.1046/j.1439-0310.2003.00849.x
Matute, D. R., Butler, I. A., Turissini, D. A., & Coyne, J. A. (2010). A test of the snowball theory for the rate of evolution of hybrid incompatibilities. Science, 329(5998), 1518–1521. https://doi.org/10.1126/science.1193440
Meier, J. I., Stelkens, R. B., Joyce, D. A., Mwaiko, S., Phiri, N., Schliewen, U. K., et al. (2019). The coincidence of ecological opportunity with hybridization explains rapid adaptive radiation in Lake Mweru cichlid fishes. Nature Communications, 10, 5391. https://doi.org/10.1038/s41467-019-13278-z
Mooney, H. A., & Cleland, E. E. (2001). The evolutionary impact of invasive species. Proceedings of the National Academy of Sciences of the United States of America, 98(10), 5446–5451. https://doi.org/10.1073/pnas.091093398
Muhlfeld, C. C., Kalinowski, S. T., McMahon, T. E., Taper, M. L., Painter, S., Leary, R. F., & Allendorf, F. W. (2009). Hybridization rapidly reduces fitness of a native trout in the wild. Biology Letters, 5(3), 328–331. https://doi.org/10.1098/rsbl.2009.0033
Nakagawa, S., & Santos, E. S. A. (2012). Methodological issues and advances in biological meta-analysis. Evolutionary Ecology, 26(5), 1253–1274. https://doi.org/10.1007/s10682-012-9555-5
Niemelä, P. T., & Dingemanse, N. J. (2014). Artificial environments and the study of “adaptive” personalities. Trends in Ecology and Evolution, 29(5), 245–247. https://doi.org/10.1016/j.tree.2014.02.007
Osborn, A., & Briffa, M. (2017). Does repeatable behaviour in the laboratory represent behaviour under natural conditions? A formal comparison in sea anemones. Animal Behaviour, 123, 197–206. https://doi.org/10.1016/j.anbehav.2016.10.036
Pagano, A., & Joly, P. (1999). Limits of the morphometric method for field identification of water frogs. Alytes, 16(3), 130–138.
Payseur, B. A. (2010). Using differential introgression in hybrid zones to identify genomic regions involved in speciation. Molecular Ecology Resources, 10(5), 806–820. https://doi.org/10.1111/j.1755-0998.2010.02883.x
Rhode, J. M., & Cruzan, M. B. (2005). Contributions of heterosis and epistasis to hybrid fitness. The American Naturalist, 166(5), E124–E139. https://doi.org/10.1086/491798
Ryan, M. E., Johnson, J. R., & Fitzpatrick, B. M. (2009). Invasive hybrid tiger salamander genotypes impact native amphibians. Proceedings of the National Academy of Sciences of the United States of America, 106(27), 11166–11171. https://doi.org/10.1073/pnas.0902252106
Sakai, A. K., Allendorf, F. W., Holt, J. S., Lodge, M., Molofsky, J., With, K. A., et al. (2001). The population biology of invasive species. Annual Review of Ecology, Evolution, and Systematics, 32, 305–332.
Schindelin, J., Rueden, C. T., Hiner, M. C., & Eliceiri, K. W. (2015). The ImageJ ecosystem: An open platform for biomedical image analysis. Molecular Reproduction & Development, 82, 518–529. https://doi.org/10.1002/mrd.22489
Schwenk, K., Brede, N., & Streit, B. (2008). Introduction. Extent, processes and evolutionary impact of interspecific hybridization in animals. Philosophical Transactions of the Royal Society B Biological Sciences, 363(1505), 2805–2811. https://doi.org/10.1098/rstb.2008.0055
Seehausen, O. (2004). Hybridization and adaptive radiation. Trends in Ecology and Evolution, 19(4), 198–207. https://doi.org/10.1016/j.tree.2004.01.003
Shik, J. Z., & Dussutour, A. (2020). Nutritional dimensions of invasive success. Trends in Ecology and Evolution, 35(8), 691–703. https://doi.org/10.1016/j.tree.2020.03.009
Šimková, A., Janáč, M., Hyršl, P., Krasnovyd, V., & Vetešník, L. (2021). Vigour-related traits and immunity in hybrids of evolutionary divergent cyprinoid species: Advantages of hybrid heterosis? Journal of Fish Biology, 98(4), 1155–1171. https://doi.org/10.1111/jfb.14648
Stelkens, R. B., Schmid, C., & Seehausen, O. (2015). Hybrid breakdown in cichlid fish. PLoS ONE, 10(5), 1–11. https://doi.org/10.1371/journal.pone.0127207
Stelkens, R., & Seehausen, O. (2009). Genetic distance between species predicts novel trait expression in their hybrids. Evolution, 63(4), 884–897. https://doi.org/10.1111/j.1558-5646.2008.00599.x
Theodosopoulos, A. N., Hund, A. K., & Taylor, S. A. (2019). Parasites and host species barriers in animal hybrid zones. Trends in Ecology and Evolution, 34(1), 19–30. https://doi.org/10.1016/j.tree.2018.09.011
Thompson, K. A., Urquhart-Cronish, M., Whitney, K. D., Rieseberg, L. H., & Schluter, D. (2021). Patterns, predictors, and consequences of dominance in hybrids. American Naturalist, 197(3), E72–E88. https://doi.org/10.1086/712603
Vanhaecke, D., de Leaniz, C. G., Gajardo, G., Young, K., Sanzana, J., Orellana, G., et al. (2012). DNA barcoding and microsatellites help species delimitation and hybrid identification in endangered galaxiid fishes. PLoS ONE, 7(3), e32939. https://doi.org/10.1371/journal.pone.0032939
Walsh, J., Maxwell, L. M., & Kovach, A. I. (2018). The role of divergent mating strategies, reproductive success, and compatibility in maintaining the Saltmarsh-Nelson’s sparrow hybrid zone. The Auk, 135(3), 693–705. https://doi.org/10.1642/AUK-17-218.1
Walsh, J., Olsen, B. J., Ruskin, K. J., Gregory Shriver, W., O’Brien, K. M., & Kovach, A. I. (2016). Extrinsic and intrinsic factors influence fitness in an avian hybrid zone. Biological Journal of the Linnean Society, 119(4), 890–903. https://doi.org/10.1111/bij.12837
Wei, X., & Zhang, J. (2018). The optimal mating distance resulting from heterosis and genetic incompatibility. Science Advances, 4(11), 1–8. https://doi.org/10.1126/sciadv.aau5518
Acknowledgements
This study was funded by the Italian Ministry for Research (PRIN project 2017KLZ3MA, Hybrind). We thank D. Canestrelli for insightful discussions. The comments of one anonymous reviewer improved an early draft of the manuscript.
Funding
Open access funding provided by Università degli Studi di Milano within the CRUI-CARE Agreement.
Author information
Authors and Affiliations
Contributions
MM and GFF conceived and designed the study and wrote the first draft of the manuscript. All authors performed the analyses and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Muraro, M., Falaschi, M. & Ficetola, G.F. Patterns of Performance Variation Between Animal Hybrids and their Parents: A Meta-analysis. Evol Biol 49, 482–496 (2022). https://doi.org/10.1007/s11692-022-09585-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11692-022-09585-x