Abstract
RELATED TO AP2.12 (RAP2.12) is one of the Ethylene Response Factors (ERF) transcription factor and plays a key role in controlling plant root bending and responding to multiple abiotic stresses including hypoxia stress. In this study, FmRAP2.12 gene was isolated and characterized from Fraxinus mandshurica Rupr. The open reading frame (ORF) of FmRAP2.12 was 1170 bp and encoded a protein of 389 amino acids. The conserved domains, three-dimensional phylogenetic relationship of FmRAP2.12 was also investigated. Quantitative real-time (qRT-PCR) analyzed the expression of FmRAP2.12 in different tissues. The expression level of FmRAP2.12 was highest in roots followed by leaves, and lowest in male flowers. Abiotic stress and hormone signal-induced expression was established using qRT-PCR. Salt stress induced FmRAP2.12 to a double peak pattern: the first peak value was at 6 h and the second at 72 h. Drought stress also induced FmRAP2.12 to a double peak pattern: the first at 6 h and the second at 48 h. FmRAP2.12 was up-regulated after initiation of gibberellic acid (GA3) treatment, with a one peak pattern at 24 h. FmRAP2.12 may not respond to cold stress and Abscisic acid (ABA) treatment. The transient overexpression of FmRAP2.12 caused the up-expression of downstream key genes of abiotic stress response and gibberellin pathway. Our research reveals the molecular characteristic and expression patterns under abiotic stress and hormone condition of FmRAP2.12, providing support for the genetic improvement of F. mandshurica at a molecular level.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
During their life cycle, plants face various abiotic stresses like drought, floods, cold, high salinity. Exposure under these stresses will lead to osmotic stress, the accumulation of reactive oxygen species (ROS), ionic imbalances, and further interrupt vital life processes and growth and development (Munns 2002; Darwish et al. 2009; Patterson et al. 2009; Park et al. 2011; Yao et al. 2016). However, plants have evolved many regulatory strategies to reduce abiotic stress and adapt to adverse environmental conditions (Hirayama and Shinozaki 2010; Golldack et al. 2014). Among these strategies, transcription factors (TFs) such as AP2/ERF, bZIP, MYB, MYC, NAC, WRKY and zinc-finger families act as mediators by regulating the expression of many downstream stress responsive genes and playing important roles in stress tolerance (Park et al. 2011; Wang et al. 2016a; Yao et al. 2016; Kumar et al. 2017).
Ethylene Response Factor VII group (ERF-VII), one of the subfamilies of AP2/ERF transcription factors, participate in growth and development processes and in stress response mechanisms in plants (Nakano et al. 2006; Park et al. 2011; Yao et al. 2016). ERF-VII proteins contain a conserved APETALA2 (AP2) domain necessary for protein-DNA interactions (Papdi et al. 2015). A highly conserved motif MCGGAI/V (N-degron) at the N-terminal allows the ERF-VII proteins to regulate protein turnover and initiate a protein degradation pathway through an oxygen sensing mechanism (Licausi et al. 2011; Gibbs et al. 2014; Papdi et al. 2015;). In Arabidopsis thaliana, the ERF-VII subfamily contains five members, RELATED TO AP2.12 (RAP2.12), RAP2.2, RAP2.3, Hypoxia Responsive ERF1 (HRE1) and HRE2 (Papdi et al. 2015). Research on ERF-VII proteins mainly focused on their function in oxygen sensing mechanisms and acted as activators of hypoxia-induced genes to regulate oxygen deprivation (Licausi et al. 2011; Gibbs et al. 2014; Papdi et al. 2015; van Dongen and Licausi 2015). In HRE1 over-expressed transgenic Arabidopsis, the tolerance to hypoxia improved, whereas in hre1hre2, a double-knockout mutant was more sensitive in hypoxia treatment than wild plants (Licausi et al. 2011). ERF-VII regulated plant cysteine oxidase enzymes (PCOs) use molecular oxygen to produce cysteine sulfinic acid (Giuntoli et al. 2017; White et al. 2017). With oxygen deficiency or hypoxia, RAP2.12 moves from the plasma membrane into the nucleus to activate the expression of anaerobic genes such as ethanol fermentation genes, pyruvate decarboxylase 1 (PDC1) and alcohol dehydrogenase 1 (ADH1), and carbohydrate degradation genes, sucrose synthase 1 (SUS1) sucrose synthase 4 (SUS4) (Licausi et al. 2011). In Arabidopsis, RAP2.2 plays an important role in reducing hypoxia stress. The overexpression of RAP2.2 improves survival of plants under oxygen deficiency, whereas in the RAP2.12 T-DNA gene knockout lines, survival rates are lower than wild type plants (Hinz et al. 2010). The ERF-VII subfamily family of proteins not only participate in regulation under hypoxia stress, but are also involved in other stress and hormone responses. In rice, submergence 1 (SUB1A), an ERF-VII subfamily member, increased the expression of genes associated with acclimation to dehydration and enhanced recovery from drought at the vegetative stage (Fukao et al. 2011). Overexpression of SUB1A also enhanced Abscisic acid (ABA) responsiveness and increased the accumulation of ROS scavenging enzymes, resulting in activating the expression of stress inducible genes and enhancing tolerance to oxidative stress (Fukao et al. 2011). In Arabidopsis, AtERF71/HRE2 responded to abiotic stress such as salt and osmotic stress (Park et al. 2011). Under oxygen deficiency, primary root curvature was controlled by ERFVII activity by mediating auxin signaling in the root tip. The bending of Arabidopsis root was inhibited by RAP2.12, whereas in rap2.12-1 seedlings, the primary root curvature was exaggerated. Overexpression of RAP2.12 will reduce the abundance of the auxin efflux carrier, pin formed 2 (PIN2) protein. Overexpression of HRE2 will inhibit root bending (Eysholdt-Derzso and Sauter 2017).
Fraxinus mandshurica grows in cool-temperate forest ecosystems and is wildly distributed in northeastern China, Korea, Japan, and eastern Russia (Kong et al. 2012; Drenkhan et al. 2014). The Manchurian ash, F. mandshurica, has excellent cold tolerance, produces a hard wood with high value and has good horticultural qualities (Looney et al. 2016; Villari et al. 2016). Previous research on Fraxinus spp. focused on disease and pest control (Showalter et al. 2018), seed germination (Zhang et al. 2015), nutritional development (Wang et al. 2018; Zhou et al. 2018), reproductive morphological differences (Zhu et al. 2016), and ecology and evolution (Li et al. 2017). With regards stress tolerance, He et al. (2016) reported that changes in the circadian clock or 24-hour rhythm affects the drought tolerance of F. mandshurica with DNA methylation variation involved in the drought response (Zeng et al. 2015). However, research around the molecular mechanisms of stress tolerance is lacking. In this study, FmRAP2.12, an ERF-VII group transcription factor, was isolated and the three-dimensional phylogenetic relationship of FmRAP2.12 was characterized. The expression profiles of FmRAP2.12 in different tissues under abiotic stress and hormone signals, and the expression of downstream genes were analyzed using qRT-RCR. This research should provide valuable information of FmRAP2.12 genes in the response to abiotic stress and hormones for tree breeding at the molecular level.
Materials and methods
Plant material and growth conditions
Fraxinus mandshurica seeds were surface sterilized and germinated at 25 °C (16-h light/8-h dark). The cultured F. mandshurica thirty-day-old seedlings was used for tissue-specific expression analysis, including the main roots, stems, leaves, and buds. Male and female flowers were collected from the Northeast Forestry Experimental Forest Farm in March. Gene cloning and expression analysis were carried out in the Department of Forest Bioengineering of the Northeast Forestry University.
Cloning and identification of the FmRAP2.12 gene
The MiniBEST Plant RNA Extraction Kit (Takara Bio, Inc., Japan) was used for total RNA extraction. The cDNA synthesized was created using the PrimeScript First Strand cDNA Synthesis Kit (Takara Bio, Inc., Japan). The putative FmRAP2.12 unigenes were identified using consensus sequences of RAP2.12 (AT1G53910) and AP2 domains of Arabidopsis for a basic, local alignment search tool (BLAST) of the F. mandshurica TSA database. FmRAP2.12 was analyzed and assembled for similarity with the DNAMAN software package. The full-length cDNA of FmRAP2.12 was obtained by polymerase chain reaction (PCR) using the primers FmRAP2.12-F (5′-ACAGCAAAAGGAAATTATCAG-3’) and FmRAP2.12-R (5′-TCAACAATGCACATGAAGGCT-3′). The FmRAP2.12 gene sequence has been submitted to GenBank with the accession number MH061003.
Sequence features and phylogenetic analysis of FmRAP2.12
The NCBI database BLAST method (https://blast.ncbi.nlm.nih.gov/Blast.cgi) searched for homologous sequences of the full-length cDNA sequence of FmRAP2.12. The physical and chemical properties of the amino acid sequence was analyzed by Protparam (http://web.expasy.org/protparam) (Gasteiger et al. 2005). The online tool TMPred was used for the prediction of transmembrane regions and orientation (https://embnet.vital-it.ch/software/TMPRED_form.html) (Hofmann 1993). The secondary structure of FmRAP2.12 was predicted by NPSA GOR4 (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_gor4.html) (Sen et al. 2005). Multiple sequence alignment was carried out using CLC Genomics Workbench. The conserved domains of FmRAP2.12 were analyzed by the NCBI Conserved Domain Database (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml). The phylogenetic tree was constructed by MEGA 5.0 with the Neighbor-Joining method and 1000 replicates of bootstrap analysis.
FmRAP2.12 protein three-dimensional structure modeling
FmRAP2.12 protein structure was not available in the protein structure databases, therefore protein sequences of FmRAP2.12 were submitted to the I-TASSER server (http://zhanglab.ccmb.med.umich.edu/ITASSER/) (Wang et al. 2016b, 2017). This tool produced a protein model based on homology modeling and threading. For homology modeling of FmRAP2.12, the PDB templates used were PDB: 2nbiA (identity 86.1%, coverage 93.9%). ModRefiner online software was further used to refine the FmRAP2.12 protein model (http://zhanglab.ccmb.med.umich.edu/ModRefiner/) (Xu and Zhang 2011).
Abiotic stresses and hormones signal treatments
Thirty-day F. mandshurica seedlings were subjected to different abiotic stresses and hormone signaling treatments. Seedlings were transferred into liquid Murashige and Skoog (MS) medium at 4 °C as a cold treatment. For salt and drought stress treatments, seedlings were transferred into liquid MS medium containing 200 mm·L−1 NaCl and 20% w/v PEG6000, respectively. For hormone treatment, seedlings were transferred into liquid MS medium containing 100 μmol/L ABA (abscisic acid) and 100 μmol/L GA3 (gibberellic acid). Whole seedlings were collected at 0, 6, 12, 24, 48 and 72 h, immediately frozen in liquid nitrogen and stored at -80 °C until RNA extraction.
Transient overexpression of FmRAP2.12 gene
The full-length cDNA of FmRAP2.12 was driven by CaMV35 s promoter and cloned into pA7-GFP expression vector as overexpression of FmRAP2.12 (OxFmRAP2.12). An empty vector pA7-GFP (35 s-GFP) was used as a control (Supplementary Fig. S1). The two vectors were transformed into Agrobacterium tumefaciens GV1301 strain. Twenty-day-old F. mandshurica wild-type (WT) seedlings were used for transient transformation, a method described for birch (Betula platyphylla Suk. by Zhang et al. (2012). Co-culturing with Agrobacterium resulted in the transient overexpression of FmRAP2.12.
Expression analysis of FmRAP2.12 gene and downstream genes
The quantified RNA was reverse-transcribed into cDNA using the PrimeScript™ RT reagent kit with gDNA Eraser (Perfect Real-Time) (Takara Bio, Inc., Japan) according to the manufacturer’s instructions. Quantitative RT-PCR (qRT-PCR) was conducted in a 7500 Real-Time PCR system (Applied Biosystems, Forster City, USA) using the Takara SYBR® Premix Ex Taq™ II (Perfect Real Time) (Takara Bio, Inc., Japan). This is a two-step thermal cycling program with the following conditions: 30 s at 94 °C for the initial denaturation, followed by 40 cycles of 5 s at 94 °C and 34 s at 60 °C, and a final melt-curve at 70–94 °C. All reactions were performed in triplicate to ensure technical and biological reproducibility, and the relative abundance of the transcripts was calculated with 7500 Software v 2.0.6 (Applied Biosystems, Forster City, USA) using the comparative 2-△△CT method (Livak and Schmittgen 2001). Tubulin was used as an internal control to determine the expression levels of the target genes. The quantitative RT-PCR (qRT-PCR) primer pairs are shown in Supplemental Table S1. The significant differences of the expression levels were analyzed using one-way ANOVAs in the SPSS version 22 software. P < 0.05 was considered statistically significant, and P < 0.01 was considered statistically extremely significant.
Results
Nucleotide sequence and protein properties of the FmRAP2.12 gene
The FmRAP2.12 full-length cDNA sequence (GenBank: MH061003) was isolated from F. mandshurica. The FmRAP2.12 CDS and protein sequences were predicted and compared (Fig. 1A). The analysis from ProtParam software showed that the ORF of FmRAP2.12 was 1170 bp and it encoded a protein of 389 amino acids with a predicted molecular mass of 42.7 kDa and a theoretical isoelectric point (pI) of 4.73. The instability index was computed to be 45.37, which classified the protein as unstable. The grand average of hydropathicity (GRAVY) was -0.688, indicating that it was a hydrophilic protein. The transmembrane prediction showed that the FmRAP2.12 protein has one possible transmembrane helix located at 17 to 35 aa (Fig. 1B). This indicates that the FmRAP2.12 protein may have transmembrane capabilities. Secondary structure analysis of the FmRAP2.12 protein revealed that FmRAP2.12 consisted of α-helix (25.7%), extended strand (15.4%) and random coil (58.9%) (Fig. 1C). The conserved domains of the FmRAP2.12 protein was further analyzed using the NCBI CD-search database (http://structure.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). The results indicated that FmRAP2.12 contained AP2 domain ranging from 122 to 179 aa and belonged to the AP2 superfamily (Fig. 1D).
The bioinformatic analysis of FmRAP2.12. aFmRAP2.12 ORF and deduced amino acid sequence are shown in Fig. 1. The start codon (ATG) is denoted as a gray box and the stop codon (TAG) is marked with an asterisk; b FmRAP2.12 helix prediction. The arrow indicates a transmembrane helix; c FmRAP2.12 protein secondary structure prediction, letter h means α-helix, letter e means extended strand, and letter c means random coil; d FmRAP2.12 protein domains prediction
Structural analysis of the FmRAP2.12 protein
To investigate the three-dimensional (3D) structure of the FmRAP2.12 protein, the sequence was submitted to the I-TASSER server. Based on homology modeling and threading, the FmRAP2.12 protein 3D model was formed. Further refinement used the ModRefiner tool (Fig. 2A, 2B). Using the COACH method, five ligand binding sites (THR154, GLU157, ALA158, ALA161 and TYR162) were then predicted, and the ligand of FmRAP2.12 was also predicted based on a PDB model (5bsaB) (Fig. 2C). The sequence of 5basB, local blastx against F. mandshurica TSA data base was used to obtain FmHistone 3.2; the homology between FmHistone 3.2 and 5basB was 77.94% (Fig. 2D), which indicated that FmHistone 3.2 may acted as a ligand and interacted with FmRAP2.12. Based on homologous gene ontology (GO) templates in PDB, GO terms were predicted for the FmRAP2.12 protein. The predicted functions were molecular functions GO:0070011 and GO:0043167 (peptidase activity and acting on L-amino acid peptides), and biological process GO:0019538 (protein metabolism).
Three-dimensional protein model of FmRAP2.12 protein, predicted by I-TASSER. a FmRAP2.12 3D structure displayed in solid ribbon style. N-to-C Terminal: colors residues in a continuous gradient from blue at the N-terminus through white to red at the C-terminus; b FmRAP2.12 3D structure displayed in solid ribbon style, secondary Type: colors according to secondary structure. Helices were red, turns were green, and coils were white; c FmRAP2.12 3D structure displayed in tube style: the AP2 domain was yellow, 5 predicted ligand binding sites (THR154, GLU157, ALA158, ALA161 and TYR162) were light blue balls; d Homologous sequence alignment between FmHistone 3.2 and 5basB
Homology analysis and phylogenetic relationship of FmRAP2.12
For comparison, Blastp and Conserved Domains Search in the NCBI database revealed that the amino acid sequence of FmRAP2.12 shared high similarities with homologous genes from other species, including Olea europaea var. sylvestris (XP_022844420.1), Populus trichocarpa (XP_002320996.1), Betula platyphylla (AJK26921.1), Sesamum indicum (XP_011072679.1), Nicotiana tabacum (NP_001312385.1), Arabidopsis thaliana (OAP16730.1), and Hevea brasiliensis (XP_021672838.1). FmRAP2.12 and seven other amino acid sequences were aligned using CLC Genomics Workbench 8 (Fig. 3). Homologous sequence alignment and combined NCBI conserved domains prediction; the results show that FmRAP2.12 and the seven other proteins shared an AP2 conserved domain (Fig. 3).
Multiple sequence alignment of FmRAP2.12, consensus sequences of different plant species (Olea europaea var. sylvestris, Populus trichocarpa, Betula platyphylla, Sesamum indicum, Nicotiana tabacum, Arabidopsis thaliana, and Hevea brasiliensis) were achieved by using CLC Genomics Workbench 10. The colored bars at the bottom represent the conservation percentage. Region of the conserved AP2 domain is framed
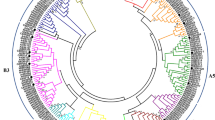
To analyze the phylogenetic relationships between FmRAP2.12 and the homologous sequences of RAP2.12 s in other plants, a phylogenetic tree was constructed using MEGA 5.0 software. FmRAP2.12 and 21 other amino acid sequences were used, including 15 dicotyledons, five monocotyledons and one bryophyte (Fig. 4). The phylogenetic tree revealed a clear boundary between the RAP2.12 proteins of dicotyledons, monocotyledons and bryophytes. FmRAP2.12 was most closely related to Oleaceae species, OeRAP2.12 (Olea europaea var. sylvestris, XP_022844420.1), Pedaliaceae species, SiRAP2.12 (Setaria italica, XP_012698581.1) and Solanaceae species, CaRAP2.12 (Capsicum annuum, XP_016562444.1), SlERF3 (Solanum lycopersicum, XP_010318409.1), NtRAP2.12 (Nicotiana tabacum, NP_001312385.1), and SiRAP2.12 (Solanum tuberosum, NP_001305599.1), as these species were grouped into a clade (Fig. 4).
Phylogenetic relationship of FmRAP2.12 proteins. RAP2.12 proteins sequences from 22 plant species were obtained from the NCBI database (http://www.ncbi.nlm.nih.gov/) using the BLASTP method. The alignment was constructed using the ClustalW method, and the phylogenetic tree was constructed using the neighbor-joining (Yao et al.) method with MEGA 5. 0 software. RAP2.12 is denoted by a red triangle. CaRAP2.12 (Capsicum annuum), SlERF3 (Solanum lycopersicum), NtRAP2.12 (Nicotiana tabacum), and SiRAP2.12 (Solanum tuberosum), FmRAP2.12 (F. mandshurica), OeRAP2.12 (Olea europaea var. sylvestris), FsERF (Fagus sylvatica), QrERF1 (Quercus robur), ZjRAP2.12 (Ziziphus jujuba), VvRAP2.12 (Vitis vinifera), PaRAP2.12 (Prunus avium), MdRAP2.12 (Malus domestica), PbRAP2.12 (Pyrus x bretschneideri), AtRAP2.12 (Arabidopsis thaliana), TaERF1 (Triticum aestivum), DvERF (Dasypyrum villosum), OsEREBP1 (Oryza sativa Japonica Group), SbERF1 (Solanum tuberosum), ZmRAP2.12 (Zea mays), PpAP2/ERF (Physcomitrella patens). (Different colors represent different clades, different phylum marked outside)
Tissue-specific expression analysis of FmRAP2.12
A real-time qRT-PCR was performed using total RNAs isolated from different tissues, including the main roots, stems, leaves, buds, male and female flowers, and seeds of F. mandshurica. The expression level of FmRAP2.12 was highest in the roots followed by the leaves, and lowest in the male flowers (Fig. 5).
Expression patterns of FmRAP2.12 gene in different tissues. The expression profile data of FmRAP2.12 gene in main root, stems, leaves, buds, male flowers, female flowers, and seeds were obtained through quantitative RT-PCR. Different letters above bars within statistically significant differences between different tissues at P < 0.05 level according to Duncan’s multiple range test
Abiotic stress and hormone signal-induced expression analysis of FmRAP2.12
F. mandshurica seedlings were exposed to cold, salt (NaCl), and drought (PEG) stress, as well as ABA and GA3 hormone signal treatments. FmRAP2.12 gene expression was induced under salt and drought conditions, but under cold was not significantly induced (Fig. 6A–E). Salt stress induced FmRAP2.12 to a double peak expression: the first was at 6 h and the second at 72 h (Fig. 6A); expression levels at the peaks were 16.6 and 16.8-fold more than at 0 h, respectively. Drought stress also induced FmRAP2.12 to a double peak expression: the first was at 6 h and the second at 48 h (Fig. 6B); the expression levels were 4.9 and 5.3-fold more than at 0 h, respectively. While the expression patterns under salt and drought conditions were similar, the induction pattern was different under cold stress. FmRAP2.12 was induced to a high value at 72 h but level of expression was only 2.35-fold than at 0 h; during the other times, expression levels were not significantly different from that at 0 h (Fig. 6C).
Quantitative assay results for FmRAP2.12 under different stress. The relative expressions of FmRAP2.12 was obtained through quantitative RT-PCR. Values are the means of three replicates, and standard errors are indicated as vertical lines on the top of each bar. Different letters above bars indicate statistically significant differences between different treatment times at P < 0.05 according to Duncan’s multiple range test
The expression patterns of FmRAP2.12 for the hormone treatments were inconsistent. FmRAP2.12 was up-regulated after initiation of the GA3 treatment with a one peak pattern at 24 h. The expression level was 12.7-fold above 0 h (Fig. 6D). In contrast, ABA induced FmRAP2.12 expression with a peak at 48 h; however, similarly to the expression under cold treatment, the expression level was only 2.56-fold than at 0 h. During the other times, expression levels were not significantly different from that at 0 h (Fig. 6E). These results indicate that FmRAP2.12 responded to salt and drought stresses and to GA3 treatments, but not responded to cold stress and ABA treatment.
Functional assay of transient overexpression of FmRAP2.12
Previous studies have shown that RAP2.12 has multiple functions and plays an important role in response to stresses such as hypoxia, salinity, osmotic stress, and response to hormones (Park et al. 2011; Papdi et al. 2015; Eysholdt-Derzso and Sauter 2017). The results of this study show that the FmRAP2.12 gene was involved in the respond to salt, drought and GA3 treatment in F. mandshurica. To study the putative genes involved in these responses, an Agrobacterium-mediated transient overexpression experiment of FmRAP2.12 in F. mandshurica seedlings was carried out (Zhang et al. 2012). The transgenic expression levels of FmRAP2.12 and the expression levels of downstream genes was determined using qRT-PCR (Fig. 7). Compared with the control (35 s-GFP) and overexpression group (OxFmRAP2.12, 35 s::FmRAP2.12-GFP), the relative expression level of FmRAP2.12 increased, which verified that the transient transformation was successful. The results show that, with the overexpression of FmRAP2.12, the gene expression levels of relative or downstream genes changed differently. FmHRE1, FmHRE2, FmRAP2.2 and FmRAP2.3, the other four members of ERF-VII subfamily genes, showed different expression patterns. The expression of FmHRE2 and FmRAP2.2 were significantly down-expressed, but FmHRE1 and FmRAP2.3 showed no difference. Notably, FmHRA1 and FmHSP18.2, (anoxia-responsive heat shock factor HSP18.2), were also significantly down-expressed, and function similarly to ERF-VII subfamily members associated with hypoxic stress response (Fig. 7). The other stress response genes, FmPDC1, FmADH1, topless-related (FmTPR2) and histone deacetylase (FmHDA1), were significantly up-expressed (Fig. 7). Similarly, with the GA3 response genes, GA insensitive (FmGAI), repressor of GA (FmRGA), FmTCP2, FmTCP15 and ethylene response 1 (FmETR1) were also significantly up-expressed (Fig. 7).
Quantitative assay results for FmRAP2.12 in F. mandshurica under different types of stress. The relative expressions of FmRAP2.12 were obtained through quantitative RT-PCR. Values are the means of three replicates and standard errors are indicated as vertical lines on the top of each bar. **statistically significant differences at P < 0.05, ***statistically extremely significant differences at P < 0.01 according to Duncan’s multiple range test
Discussion
Transcription factors (TFs) play important roles in regulating of different genes to the response to stress and in coordinating developmental regulation (Mizoi et al. 2012; Rushton et al. 2012). Plant-specific ERF family proteins are well- known to improve the tolerance to abiotic stresses such as high salinity, cold, drought, disease or excess water, and to adapt to various environmental conditions (Mizoi et al. 2012; Rehman and Mahmood 2015; Thirugnanasambantham et al. 2015). The best-known ERF family members are Dehydration Responsive Element Binding1 (DREB1/CBF) and DREB2 genes (Sakuma et al. 2002). In research on different plant species, the DREB1/CBF genes mainly participated cold response whereas the DREB2 genes were generally responsive to excess water or heat stress (Zhao et al. 2012; Artlip et al. 2013; Girardi et al. 2013). SUB1A, one of the ERF-VII TFs, increased osmotic and oxidative stress tolerances in rice (Fukao et al. 2011), and the ectopic expression of HvRaf in Arabidopsis enhanced tolerant towards salt and pathogenic fungi (Jung et al. 2007).
In this study, we isolated and characterized the FmRAP2.12 gene, one of the ERF-VII family members, from F. mandshurica. Bioinformatic analysis showed that the FmRAP2.12 revealed a 1170 bp ORF, encoding a protein of 389 amino acids. A conserved domains search showed the FmRAP2.12 protein contained an AP2 domain ranging from 122 to 179 AA and belonged to AP2 superfamily (Fig. 1A–D). By using I-TASSER online software and PDB model database, we built a three-dimensional model of FmRAP2.12. We predicted one of the possible binding ligands of the protein was FmHistone 3.2, indicating that FmRAP2.12 may interacted with FmHistone 3.2 to act as a regulator under stress conditions. Homology analysis showed that the AP2 domain structure is conserved in FmRAP2.12 and in other plant species, which confirms the prediction of NCBI conserved domains. It is well- known that AP2 subfamily proteins contain one or two AP2/ERF domains and are involved in the regulation of developmental processes and stresses responses (Boutilier et al. 2002; Cao et al. 2015). The RAV, (Related to ABI3/VP1) proteins, contains one AP2/ERF DNA-binding domains and B3 DNA-binding domains which could be regulated by brassinosteroids and ethylene, and participate in biotic and abiotic stress responses (Cao et al. 2015; Lim et al. 2015). Combined with the results of the 3D structure modeling analysis of FmRAP2.12 protein, it was found that the ligand binding sites were also located in the AP2 domain, indicating that the domain may play a key role in the interaction and regulation of FmRAP2.12 with other proteins.
In this research, FmRAP2.12 gene expression patterns in different tissues were expressed significantly higher in the roots than in other organs (Fig. 5). In response to abiotic stress and hormone treatments, the expression patterns of FmRAP2.12 under salt and drought stress were similar, revealing double peak expression patterns (Fig. 6A–B). However, the expression pattern of FmRAP2.12 under GA3 treatment revealed a one- peak pattern (Fig. 6D). The expression level of FmRAP2.12 under cold stress and ABA treatment showed almost no significant changes (Fig. 6C and E). The cause of this may likely be related to the treatment time, treatment intensity or hormone concentration, and requires further research. We concluded that FmRAP2.12 gene responded to salt and drought abiotic stresses and GA3 treatments but not responded to cold stress and ABA treatment (Fig. 6A–E). Moreover, overexpressing FmRAP2.12 caused significant changes in the expression of a series of key genes involved in abiotic stress response and GA3 pathways. FmHRE1 and FmHRE2, two members of the ERF-VII subfamily, showed significant down-expression. The other two genes, FmHRA1 and FmHSP18.2, which related to the ERF-VII subfamily and associated with hypoxic stress response, were also significantly down-expressed (Fig. 7). Whereas the other abiotic stress response genes, such as FmADH1, FmPDC1, FmTPR2 and FmHDA1, and GA3 response genes such as DELLA family members FmGAI and FmRGA1, showed significantly up-expression (Fig. 7). Previous studies have shown that the RAP2.12 protein could activate the expression of ADH1 and PDC1, and the overexpressing of AtADH1 and AtPDC1 improved the resistance to salt, drought and other abiotic stresses in Arabidopsis. (Licausi et al. 2011; Rasheed et al. 2018; Shi et al. 2017). In rice, IDS1 (Indeterminate Spikelet 1), an AP2 type transcription factor, could interact with TPR1 and HDA1 to form IDS1-TPR1-HDA1, a transcriptional repression complex, and regulate salt tolerance under stress conditions (Licausi et al. 2010). However, the ERF VII family also cross talk with hormone signaling pathways. Yeast two-hybrid screen of Arabidopsis identified RAP2.3 and RAP2.12 which could interact with the DELLA protein GAI (Marín-de la Rosa et al. 2014). It is well- known that RGA and GAI have a major role in salt-induced plant growth regulation and confer on Arabidopsis the capacity of salt tolerance (Achard et al. 2006; Marín-de la Rosa et al. 2014). All the gene expression evidence of RAP2.12 is similar to our results and indicates that RAP2.12 plays an important role in regulating plant response to stresses and hormone signal responses.
The FmRAP2.12 gene was mainly expressed in the roots in F. mandshurica. Moreover, FmRAP2.12 significantly responded to salt and drought stress and to GA3 treatment. Plant roots are sensitive to abiotic stress and hormonal signals. The expression of FmRAP2.12 may contribute to the resistance by plants to these stresses. Protein 3D structure modeling predicted that the potential protein binding ability of AP2 domain conferred on FmRAP2.12 the capacity to regulate downstream genes. Thus, on the one hand, FmRAP2.12 regulated the expression of a series of abiotic stress response genes, such as FmADH1, FmPDC1, FmTPR2 and FmHDA1. On the other hand, FmRAP2.12 interacts with GA3 pathway genes, such as DELLA family members FmGAI and FmRGA, and indirectly respond to stress through the GA3 pathway. This study reports the gene isolation, bioinformatics analysis, tissue-specificity expression, and abiotic stress and hormone signal response of FmRAP2.12 from F. mandshurica. It provides a candidate gene for stress tolerance and supports the genetic improvement of F. mandshurica at the molecular level.
References
Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng J, Harberd NP (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science 311:91–94
Artlip TS, Wisniewski ME, Bassett CL, Norelli JL (2013) CBF gene expression in peach leaf and bark tissues is gated by a circadian clock. Tree Physiol 33:866–877
Boutilier K, Offringa R, Sharma VK, Kieft H, Ouellet T, Zhang L, Hattori J, Liu C-M, van Lammeren AA, Miki BL (2002) Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14:1737–1749
Cao P, Azar S, SanClemente H, Mounet F, Dunand C, Marque G, Marque C, Teulières C (2015) Genome-wide analysis of the AP2/ERF family in Eucalyptus grandis: an intriguing over-representation of stress-responsive DREB1/CBF genes. PLoS ONE 10:e0121041
Darwish E, Testerink C, Khalil M, El-Shihy O, Munnik T (2009) Phospholipid signaling responses in salt-stressed rice leaves. Plant Cell Physiol 50:986–997
Drenkhan R, Sander H, Hanso M (2014) Introduction of Mandshurian ash (Fraxinus mandshurica Rupr.) to Estonia: is it related to the current epidemic on European ash (F. excelsior L.)? Eur J Forest Res 133:769–781
Eysholdt-Derzso E, Sauter M (2017) Root bending is antagonistically affected by hypoxia and ERF-mediated transcription via auxin signaling. Plant Physiol 175:00555–02017
Fukao T, Yeung E, Bailey-Serres J (2011) The submergence tolerance regulator SUB1A mediates crosstalk between submergence and drought tolerance in rice. Plant Cell 110:080325
Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins M, Appel R, Bairoch A, Walker J (2005) Protein identification and analysis tools on the ExPASy server. The proteomics protocols handbook. Humana Press, Humana, pp 571–607
Gibbs DJ, Isa NM, Movahedi M, Lozano-Juste J, Mendiondo GM, Berckhan S, Marín-de la Rosa N, Conde JV, Correia CS, Pearce SP (2014) Nitric oxide sensing in plants is mediated by proteolytic control of group VII ERF transcription factors. Mol Cell 53:369–379
Girardi CL, Rombaldi CV, Dal Cero J, Nobile PM, Laurens F, Bouzayen M, Quecini V (2013) Genome-wide analysis of the AP2/ERF superfamily in apple and transcriptional evidence of ERF involvement in scab pathogenesis. Sci Hortic (Amsterdam) 151:112–121
Giuntoli B, Shukla V, Maggiorelli F, Giorgi FM, Lombardi L, Perata P, Licausi F (2017) Age-dependent regulation of ERF-VII transcription factor activity in Arabidopsis thaliana. Plant, Cell Environ 40:2333–2346
Golldack D, Li C, Mohan H, Probst N (2014) Tolerance to drought and salt stress in plants: unraveling the signaling networks. Front Plant Sci 5:151
He Z, Zhan Y, Zeng F, Zhao X, Wang X (2016) Drought physiology and gene expression characteristics of Fraxinus interspecific hybrids. Plant Growth Regul 78:179–193
Hinz M, Wilson IW, Yang J, Buerstenbinder K, Llewellyn D, Dennis ES, Sauter M, Dolferus R (2010) Arabidopsis RAP2. 2: an ethylene response transcription factor that is important for hypoxia survival. Plant Physiol 153:757–772
Hirayama T, Shinozaki K (2010) Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J 61:1041–1052
Hofmann K (1993) TMbase-A database of membrane spanning proteins segments. Biol Chem Hoppe Seyler 374:166
Jung J, Won SY, Suh SC, Kim H, Wing R, Jeong Y, Hwang I, Kim M (2007) The barley ERF-type transcription factor HvRAF confers enhanced pathogen resistance and salt tolerance in Arabidopsis. Planta 225:575–588
Kong DM, Preece JE, Shen HL (2012) Somatic embryogenesis in immature cotyledons of Manchurian ash (Fraxinus mandshurica Rupr.). Plant Cell, Tissue Organ Cult 108:485–492
Kumar M, Choi J, An G, Kim SR (2017) Ectopic expression of OsSta2 enhances salt stress tolerance in rice. Front Plant Sci 8:316
Li Y, Wang X, Wang C, Liu F, Zhang Q (2017) Spatial variation in diurnal courses of stem temperature of Betula platyphylla and Fraxinus mandshurica and its influencing factors. Ying Yong Sheng Tai Xue Bao 28:3197–3207
Licausi F, Van Dongen JT, Giuntoli B, Novi G, Santaniello A, Geigenberger P, Perata P (2010) HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana. Plant J 62:302–315
Licausi F, Kosmacz M, Weits DA, Giuntoli B, Giorgi FM, Voesenek LA, Perata P, van Dongen JT (2011) Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 479:419
Lim CW, Baek W, Lim S, Han SW, Lee SC (2015) Expression and functional roles of the pepper pathogen–induced bZIP Transcription factor CabZIP2 in enhanced disease resistance to bacterial pathogen infection. Mol Plant Microbe Interact 28:825–833
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Looney CE, D’Amato AW, Palik BJ, Slesak RA (2016) Canopy treatment influences growth of replacement tree species in Fraxinus nigra forests threatened by the emerald ash borer in Minnesota, USA. Can J For Res 47:183–192
Marín-de la Rosa N, Sotillo B, Miskolczi P, Gibbs DJ, Vicente J, Carbonero P, Oñate-Sánchez L, Holdsworth MJ, Bhalerao R, Alabadí D (2014) Large-scale identification of gibberellin-related transcription factors defines Group VII ERFs as functional DELLA partners. Plant Physiol 114:244723
Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2012) AP2/ERF family transcription factors in plant abiotic stress responses. BBA-Gene Regul Mech 1819:86–96
Munns R (2002) Comparative physiology of salt and water stress. Plant, Cell Environ 25:239–250
Nakano T, Suzuki K, Fujimura T, Shinshi H (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140:411–432
Papdi C, Pérez-Salamó I, Joseph MP, Giuntoli B, Bögre L, Koncz C, Szabados L (2015) The low oxygen, oxidative and osmotic stress responses synergistically act through the ethylene response factor VII genes RAP 2.12, RAP 2.2 and RAP 2.3. Plant J 82:772–784
Park HY, Seok HY, Woo DH, Lee SY, Tarte VN, Lee EH, Lee CH, Moon YH (2011) AtERF71/HRE2 transcription factor mediates osmotic stress response as well as hypoxia response in Arabidopsis. Biochem Bioph Res Co 414:135–141
Patterson JH, Newbigin E, Tester M, Bacic A, Roessner U (2009) Metabolic responses to salt stress of barley (Hordeum vulgare L.) cultivars, Sahara and Clipper, which differ in salinity tolerance. J Exp Bot 60:4089–4103
Rasheed S, Bashir K, Kim JM, Ando M, Tanaka M, Seki M (2018) The modulation of acetic acid pathway genes in Arabidopsis improves survival under drought stress. Sci Rep 8:7831
Rehman S, Mahmood T (2015) Functional role of DREB and ERF transcription factors: regulating stress-responsive network in plants. Acta Physiol Plant 37:178
Rushton DL, Tripathi P, Rabara RC, Lin J, Ringler P, Boken AK, Langum TJ, Smidt L, Boomsma DD, Emme NJ (2012) WRKY transcription factors: key components in abscisic acid signalling. Plant Biotechnol J 10:2–11
Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration-and cold-inducible gene expression. Biochem Biophys Res Commun 290:998–1009
Sen TZ, Jernigan RL, Garnier J, Kloczkowski A (2005) GOR V server for protein secondary structure prediction. Bioinformatics 21:2787–2788
Shi HT, Liu W, Yao Y, Wei YX, Chan ZL (2017) Alcohol dehydrogenase 1 (ADH1) confers both abiotic and biotic stress resistance in Arabidopsis. Plant Sci 262:24–31
Showalter DN, Villari C, Herms DA, Bonello P (2018) Drought stress increased survival and development of emerald ash borer larvae on coevolved M anchurian ash and implicates phloem-based traits in resistance. Agr Forest Entomol 20:170–179
Thirugnanasambantham K, Durairaj S, Saravanan S, Karikalan K, Muralidaran S, Islam VIH (2015) Role of ethylene response transcription factor (ERF) and its regulation in response to stress encountered by plants. Plant Mol Biol Rep 33:347–357
van Dongen JT, Licausi F (2015) Oxygen sensing and signaling. Annu Rev Plant Biol 66:345–367
Villari C, Herms DA, Whitehill JG, Cipollini D, Bonello P (2016) Progress and gaps in understanding mechanisms of ash tree resistance to emerald ash borer, a model for wood-boring insects that kill angiosperms. New Phytol 209:63–79
Wang H, Wang H, Shao H, Tang X (2016a) Recent advances in utilizing transcription factors to improve plant abiotic stress tolerance by transgenic technology. Front Plant Sci 7:67
Wang Y, Virtanen J, Xue Z, Tesmer JJ, Zhang Y (2016b) Using iterative fragment assembly and progressive sequence truncation to facilitate phasing and crystal structure determination of distantly related proteins. Acta Crystallogr D 72:616–628
Wang Y, Virtanen J, Xue Z, Zhang Y (2017) I-TASSER-MR: automated molecular replacement for distant-homology proteins using iterative fragment assembly and progressive sequence truncation. Nucleic Acids Res 45:429–434
Wang M, Zhang WW, Li N, Liu YY, Zheng XB, Hao GY (2018) Photosynthesis and growth responses of Fraxinus mandshurica Rupr. seedlings to a gradient of simulated nitrogen deposition. Ann Forest Sci 75:1123
White MD, Klecker M, Hopkinson RJ, Weits DA, Mueller C, Naumann C, O’Neill R, Wickens J, Yang J, Brooks-Bartlett JC (2017) Plant cysteine oxidases are dioxygenases that directly enable arginyl transferase-catalysed arginylation of N-end rule targets. Nat Commun 8:14690
Xu D, Zhang Y (2011) Improving the physical realism and structural accuracy of protein models by a two-step atomic-level energy minimization. Biophys J 101:2525–2534
Yao W, Wang S, Zhou B, Jiang T (2016) Transgenic poplar overexpressing the endogenous transcription factor ERF76 gene improves salinity tolerance. Tree Physiol 36:896–908
Zeng FS, Li LL, Liang NS, Wang X, Li X, Zhan YG (2015) Salt tolerance and alterations in cytosine methylation in the interspecific hybrids of Fraxinus velutina and Fraxinus mandshurica. Euphytica 205:721–737
Zhang Y, Wang Y, Wang C (2012) Gene overexpression and gene silencing in Birch using an Agrobacterium-mediated transient expression system. Mol Biol Rep 39:5537–5541
Zhang P, Liu D, Shen H, Li Y, Nie Y (2015) Proteome analysis of dormancy-released seeds of Fraxinus mandshurica Rupr. in response to re-dehydration under different conditions. Int J Mol Sci 16:4713–4730
Zhao T, Liang D, Wang P, Liu J, Ma F (2012) Genome-wide analysis and expression profiling of the DREB transcription factor gene family in Malus under abiotic stress. Mol Genet Genomics 287:423–436
Zhou Y, He Y, Sun J, Zhang J, Zhan Y (2018) Effects of nitrogen deficiency on physiology and growth of Fraxinus mandshurica. Pak J Bot 50:179–187
Zhu Z, Qi F, Yan C, Zhan Y (2016) Sexually different morphological, physiological and molecular responses of Fraxinus mandshurica flowers to floral development and chilling stress. Plant Physiol Bioch 99:97–107
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Project funding: This work was supported by the National Key Research and Development Project of China (No. 2017YFD0600605-01) and the National Natural Science Foundation of China (NSFC) (No. 31270697).
The online version is available at http://www.springerlink.com.
Corresponding editor: Yu Lei.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Liang, N., Yu, L., Liu, C. et al. Molecular cloning and expression under abiotic stresses and hormones of the ethylene response factor VII gene FmRAP2.12 from Fraxinus mandshurica. J. For. Res. 30, 1289–1300 (2019). https://doi.org/10.1007/s11676-019-00979-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11676-019-00979-w