Abstract
The symbiotic octocoral Sinularia flexibilis is a producer of potential pharmaceuticals. Sustainable mass production of these corals as a source of such compounds demands innovative approaches, including coral cell culture. We studied various cell dissociation methodologies and the feasibility of cultivation of S. flexibilis cells on different media and cell dissociation methodologies. Mechanical dissociation of coral tissue always yielded the highest number of cells and allowed subsequent cellular growth in all treatments. The best results from chemical dissociation reagents were found with trypsin-ethylene diamine tetraacetic acid. Coral cells obtained from spontaneous dissociation did not grow. Light intensity was found to be important for coral cell culture showing an enduring symbiosis between the cultured cells and their intracellular algae. The Grace’s insect medium and Grace’s modified insect medium were found to be superior substrates. To confirm the similarity of the cultured cells and those in the coral tissue, a molecular test with Internal Transcribed Spacer primers was performed. Thereby, the presence of similar cells of both the coral cells and zooxanthella in different culture media was confirmed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soft corals are a promising source of cytotoxic agents as anticancer, anti-inflammatory, and analgesic compounds (Faith et al. 1997; Ata et al. 2003; Duque et al. 2004). The soft coral Sinularia flexibilis (family Alcyoniidae) harbors symbiotic dinoflagellate algae (zooxanthellae) and produces bioactive compounds with cytotoxic, antimicrobial, and anti-inflammatory activities (Dhu et al. 1998; Kamel and Slattery 2005; Radhika et al. 2005; Chao et al. 2006; Yu et al. 2006). The increasing interest in these resources for drug development raises a conservation concern (Hunt and Vincent 2006). To avoid overexploitation of coral ecosystems, the production of the source animal or its cells is important.
Among the approaches for sustainable exploitation is the coral cell culture with applications in fields such as ecotoxicology and biotechnology (Rodgers 2002). Techniques for cnidarian cell culture (including corals) have not been developed yet. Problems were encountered with cell dissociation, culture contamination, and medium (Frank et al. 1994; Odintsova et al. 1994; Rinkevich 1999). Due to the lack of validated aquatic invertebrate cell lines, primary cell or tissue cultures for toxicology have been used (Reviewed by Villena 2003).
Cell cultures from a gorgonian octocoral scleroblasts were maintained for more than 4 mo (Kingsley et al. 1987). A review by Rinkevich (1999) indicates that between 1988 and 1998, only three studies on coral cell culture were published (Gates et al. 1992; Frank et al. 1994). In most cases, the cells did not survive more than 1 mo. Frank et al. (1994) observed that cryopreserved cell cultures after approximately 1 yr were excluded by eukaryotic unicellular organisms. Again, between 1998–2004, there were no successful studies published on long term cnidarian cell cultures (Frank and Rinkevich 1999; Kopecky and Ostrander 1999; Schmid et al. 1999; Domart-Coulon et al. 2001, 2004) even though some encouraging reports were presented (reviewed by Rinkevich 2005).
The failure to establish long-lasting and proliferating cell cultures has been attributed to the lack of vital information regarding cell requirements and their physiology and biochemical patterns in vitro, improper comparisons between vertebrate–invertebrate cell requirements (Rinkevich 1999), as well as insufficient information about the classification and identification of cell types (Puverel et al. 2005). To extend the functional survival of coral cells, cell culture protocols have to be improved (Domart-Coulon et al. 2004).
The principal aim of this study, therefore, was to investigate the establishment of long-lasting cell culture from the symbiotic soft coral S. flexibilis. Because both substrate composition and cell dissociation methods are critical in the survival and prolongation of cells (Frank et al. 1994; Odintsova et al. 1994; Rinkevich 1999), we examined various media and cell-dissociation methodologies. Eukaryotic endosymbionts might develop in invertebrate cell cultures instead of the target coral cells. Hence, specific tests such as genetic probes should be used to verify the nature of the cultured cells (Villena 2003). Therefore, we have also used for the first time a genetic approach to identify coral cells in culture.
Materials and Methods
Coral cell culture.
All cultivation procedures (in four stages) were executed with sterile materials; protocols were based on Gates et al. (1992), Frank et al. (1994), Kopecky and Ostrander (1999), and Hurton et al. (2005).
Organism.
Colonies of S. flexibilis were obtained from Burgers’ Zoo, Arnhem, The Netherlands and were maintained in laboratory tanks (25 ± 0.5°C, 12 h light–12 h dark period, 34‰ salinity, and a pH of 8 ± 0.5). Prior to dissociation, corals were rinsed at least ten times with sterile sea water (SSW; Instant Ocean Reef Crystals, Aquarium Systems, Sarrebourg, France) and sliced into 1-mm3 pieces with a scalpel.
Tissue Dissociation - Spontaneous.
Coral samples (1 mm2) were incubated in a 1% gentamycin–streptomycin (1:1) solution in SSW at room temperature during 4 h and transferred subsequently to the respective culture medium (see “Culture Media and Conditions”).
Mechanical dissociation.
Coral samples (1 mm2) were rinsed with SSW and cut into small pieces using scalpel. Then, the samples were rinsed again and placed in a 100-μm nylon cell strainer (BD Falcon, San Jose, CA, USA) and sieved by pressing them with a loose-fit plastic piston. Sieved cells were resuspended in 1.5 ml of SSW in 2-ml sterile eppendorf tubes. Cell suspensions were centrifuged (300×g, 10 min); the supernatant was discarded and the pellet resuspended in 1.5 ml 1% gentamycin–streptomycin solution (1:1) in SSW. This procedure was performed four times.
Chemical dissociation.
Following rinsing, coral pieces (1 mm3) were placed into eppendorf tubes (2 ml) containing one of the following dissociation reagents: trypsin-ethylene diamine tetraacetic acid (EDTA) 0.05% in SSW, pronase 0.1% in SSW or collagenase 0.05% in SSW and then shaken at 200 rpm for 40 min, 60 min, and 2 h, respectively. Cell suspensions were centrifuged and resuspended as explained for mechanical dissociation.
Culture Media and Conditions - Media.
Four enriched media were used for cultivation: Dulbecco’s modified Eagle medium (DMEM), medium 199 (Frank et al. 1994), Grace’s insect medium (GIM) and Grace’s modified insect medium (GMIM; Hurton et al. 2005). To avoid crystallization, supplements were not added (Frank et al. 1994). All media contained a 1% antibiotic cocktail (streptomycin–gentamycin 1:1). Cells in sterile sea water were used as a control.
Culture containers.
After initial attempts with 25 cm3 tissue culture flasks (T-flasks), cells were cultivated in 24-well plates with 1.5 ml of medium and 1% antibiotic cocktail (streptomycin–gentamycin 1:1).
Incubation conditions and culture maintenance.
Cultures were preserved in a climate cabinet (Snijders Scientific) with a constant temperature of 24° C and 5% (v/v) of CO2 in air. Fluorescent light (80–100 μE m−2 s−1) was applied in a 12 h light–12 h dark photoperiod. Cultures were checked daily, and culture media were refilled almost once per week or when the media were below 0.7 ml.
Culture Analysis - Growth.
Cultures were counted two times per week with an inverted microscope (Olympus CK40); pictures of all the treatments were taken using a digital camera (Olympus C-3030) with a lens (Olympus U-PMTVC) and the software AnaliSYS for photo extracting and scaling (Soft Imaging System GmbH). Three different counting methods were used: after the dissociation process and in the first stages of the experiment, counts were made by placing a graph paper transparency and counting the cells within a 1 mm2. After appearance of cellular aggregates, the total area occupied by the cells was calculated with AnaliSYS software. However, due to the low accuracy of this method in the end, counts were made based on image analysis using the software Image-Proplus for windows (version 4.5.0.29, Media cybernetics Inc.) with its manual tag tool.
Cell identification.
To identify the cells present in the cultures, two approaches were used: histology slides and genetic test.
Histology slides.
A colony fragment of 3 mm2 was rinsed in SSW at least five times and placed in a container with 1% agarose and 1% sugar in SSW. Subsequently, the sample was placed in a metal container and frozen with liquid nitrogen. Sections of 7 µm were made using a cryostat (type 2800 frigocut Reichert Jung), spread on slides, and fixated with 5 ml formaldehyde 40% neutral, 15 ml saturated picric acid, and 1 ml acetic acid 100% during 10 min. Slides were then stained with hematoxylin and eosin (3 min each).
Genetic test using Internal Transcribed Spacer primers - DNA isolation.
Coral DNA was extracted from both coral tissue (animal) and from coral cell cultures on GMIM, GIM, and DMEM media. Either a piece of tissue or some cultured cells were placed in Eppendorf tubes together with five small glass beads. The tubes were frozen using liquid nitrogen and shaken in a bead-beater for 10 s. Subsequently, five extra small glass beads were added, and the samples were frozen and shaken again for 10 s. Samples were then vortexed with 1 ml of LETS buffer (0.1 M LiCl, 0.01 M EDTA, 0.01 M Tris-HCl (pH 7.4), 0.2% sodium dodecyl sulfate) and centrifuged for 5 min at 13,000 rpm. Of the supernatant, 700 μl was transferred into a fresh 2 ml Eppendorf tube and 5 μl of proteinase K (stock 20 μg/μl was kept at −20°C) were added; samples were incubated for 1 h at 37°C. Next, 300 μl phenol and 300 μl SEVAG were added, and the samples were well vortexed and after that centrifuged for 15 min at 13,000 rpm; 600 μl of the supernatant were transferred to a clean 1.5-ml Eppendorf tube and 300 μl of SEVAG was added. Samples were mixed again and centrifuged for 15 min at 13,000 rpm; subsequently, 500 μl of the supernatant was transferred to a clean 1.5-ml Eppendorf tube and 300 μl of isopropanol (0.6 × volume) added and samples were stored for 15 min at −20°C. Tubes were centrifuged once more for 15 min at 13,000 rpm, the supernatant was discarded while the pellet was washed with 100 μl of ice-cold 70% ethanol, and samples were centrifuged for 1 min at 13,000 rpm. Then, the supernatant was discarded and the pellet dried under a vacuum. Finally, the pellet was dissolved in 50 μl of Milli-Q.
Polymerase chain reaction (PCR).
The Internal Transcribed Spacer (ITS) of the coral animal was amplified using two different primer sets: the first set consisted of primers ITS 1s (5′-GGT ACC CTT TGT ACA CAC CGC CCG TCG CT-3′) and ITS 2ss (5′-GCT TTG GGC TGC AGT CCC AAG CAA CCC GAC TC-3′; McFadden et al. 2001); the primers belonging to the second set were ITS a (5′-GGG ATC CGT TTC CGT AGG TGA ACC TGC-3′) and ITS b (5′-GGG ATC CAT ATG CTT AAG TTC AGC GGG T-3′; Coleman et al. 1994). The PCR protocol consisted of an initial denaturating step of 4 min at 94°C followed by 35 cycles of denaturation at 94°C (30 s), annealing at 52°C (60 s), elongation at 72°C (90 s), and in the end, a final elongation step at 72°C (10 min; McFadden et al. 2001). PCR reactions were performed in 25 ml volumes, using four DNA concentrations: undiluted and 10, 100, and 1,000 times diluted. Five microliters of a PCR reaction was run together with an undiluted sample for each culture medium and also for the coral (animal) in a 0.6% agarose electrophoresis gel at 70 v/cm/s; the size of the fragments was established using the marker Orange Ruler 100 bp DNA ladder.
Results and Discussion
Dissociation.
Three different dissociation techniques to isolate free cells from coral tissue were tested (Table 1). Spontaneous dissociation yielded the lowest number of free cells. The low yield, in comparison to the other techniques, could be because of the absence of extensive mechanical procedures or enzymatic reactions to separate the tissue by pressure or chemicals. In addition, for spontaneous dissociation, more washing steps are used; possibly, some of the separated cells are lost in this process. With mechanical dissociation, the highest number of free cells was obtained, which is consistent with Frank et al. (1994).
Three chemical dissociation agents were used, of which trypsin-EDTA was the most effective. Pronase was used only in two of the extractions because it caused a lot of damage to the cells, and consequently, low numbers of free cells were obtained. Pronase is a nonspecific protease that hydrolyses proteins to individual amino acids and can lead to damage of the cellular membranes (Roche Applied Science 2006). The cell yield after treatment with trypsin-EDTA was much higher. Trypsin-EDTA cleaves the proteins into peptides, hence, detaches the tissue into cell clusters, leaving more cells intact (Sigma–Aldrich 2007). Collagenase treatment did not lead to positive results in our studies. This is in contradiction with the studies of Frank et al. (1994) who found that collagenase was less harmful than trypsin-EDTA.
Growth of the coral cultured cells.
After initial cultivation experiments with T-flasks, 24-well plates were used for cultivation. In well plates, we obtained higher cell densities, and counting of cells was easier. An essential factor for cellular growth is that relatively high cell densities are used (Freshney 1994; Verkerk et al. 2007). Results of 24-well-plate growth are displayed in Fig. 1.
During the first week of cell culturing (24-well plate), viable cell numbers decreased in all media, probably due to the stress caused by the dissociation process (Freshney 1994). On day 12, the light intensity was changed from 130 to 80–100 μE m−2 s−1. After 14 d, a significant increase in the number of mechanically dissociated cells in GIM medium (M-GIM) was observed (Fig. 1). Cellular proliferation was also observed for mechanically dissociated cells grown in GMIM medium (M-GMIM) after 20 d of cultivation. Apparently, the coral cells and their intracellular zooxanthellae responded positively to decreased light intensity, indicating that they maintain a strong symbiotic relationship even after tissue dissociation. A higher cell survival time (not growth) in light than in dark was reported previously for coral cells (Kopecky and Ostrander 1999).
A slight growth of chemically dissociated cells in GMIM medium (C-GMIM) was observed on day 26. The rest of the wells, even with a low cell density, remained stable and in good conditions. DMEM and M199 were previously used in coral cell culture by Frank et al. (1994), who also tested Leibovits L-15 medium. However, the article’s discussion is based on the last mentioned media and claims that coral cells could be maintained during 1 yr using it. Data on DMEM and M199 performance were not shown, even though cellular growth was also reported. Based on the observations during that study, DMEM and M199 did not probably lead to better results than L-15 media. Kopecky and Ostrander (1999) also used DMEM as the only culture medium; nevertheless, they just investigated the time that DMEM was able to keep the cells alive without changing the medium, establishing it up to 300 h.
On the other hand, the GIM and GMIM media that performed best in the present study have not been tested in coral cell culture before. They were used by Hurton et al. (2005) for primary cultures of amebocytes of the horseshoe crab Limulus polyphemus. In comparison, they had better results with GIM and GMIM than with L-15 media.
After the rather promising cell growth on GIM and GMIM media, a less time-consuming counting technique was used. It was not possible to use a counting chamber because the cells attached to the bottom of the wells. We determined the area occupied by the cells or cell aggregates instead (Table 2). With this method, growth could clearly be determined although the accuracy needs further improvement. In addition, the cells were partially spread through the well after the medium was changed, making the area monitoring impossible.
In order to find out if the lack of cell growth in the wells containing DMEM and M199 was due to the media, new wells were inoculated from the proliferating cultures (GIM and GMIM) and grown on fresh media (GIM, GMIM, M199, and DMEM). Growth of these new cultures was evident from a series of general pictures at low magnification (×4, Fig. 2). Again, cells did not grow on media M199 and DMEM and grew on GIM and GMIM (images from media M199 and DMEM are not shown).
Picture monitoring (×4 magnification) on the cells of S. flexibilis from the inoculation 24-well plate using Image-proplus software, showing images from the proliferating cultures (GIM and GMIM) in the media GIM and GMIM (Fig. 2).
The number of cells in these general pictures was counted using the software Image-Proplus with its manual tag tool (Fig. 2) as a reliable counting methodology, based on high-quality pictures and specific well plates for microscopy. Because of the presence of other particles in the coral cell cultures, the manual tag tool was employed. An increase in the number of cells can be seen from day 2 until day 8. During the next 2 d, a decline of the number of cells is noticed in both GM-GIM and GM-GMIM wells, probably due to an overestimation in the preceding counts. Because the quality of the pictures employed during this stage was suboptimal, these low-quality pictures may be used as a visual estimation. This inoculation experiment showed the efficient performance of media GIM and GMIM. Therefore, other growth inhibitors in the wells or in the media of the remaining treatments were rejected.
From the first day of inoculation, a depletion in the number of viable cells in GIM wells for both the GM-GIM and TM-GIM was observed (Fig. 2) that was caused by a microbial contamination. This is in agreement with Frank et al. (1994), who stated that microbial contamination may result in a rapid loss of over 80% of the coral cells. After day 16, a growth interruption, and also a decrease in the number of viable cells on GIM media, was observed due to a bacterial contamination and also a failure in the light manager device (the last picture on day 30, Fig. 2); the cultures were unintentionally exposed to permanent light. Once more, due to the photosynthesizing symbiont (zooxanthellae), the importance of lighting period was observed through a decrease in the growth rates afterwards.
Cell Identification.
Because eukaryotic endosymbionts might develop in cell cultures (Pomponi et al. 1997) it is necessary to identify the target coral cells. Identification of cnidarian cell types based on morphology alone has proven to be very difficult (Puverel et al. 2005), mainly because cells adapt their shape to their endosymbionts. Thus, if a coral cell possesses a single zooxanthellae, it will be round and approximately 10 µm in diameter, but if it contains two or more alga, the host diameter will increase and its cell shape will conform to the shape in which the symbionts cluster (Gates et al. 1992). On the other hand, coral cells are colourless while their photosynthetic endosymbionts, contain multiple pigments (Smith 2001). This can help distinguish the cells by appearance.
Histological sections.
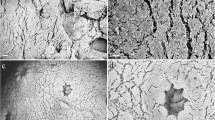
To verify that the cultured cells were in fact zooxanthellae-containing coral cells, three histological sections were prepared from coral tissue, analyzed microscopically, and compared to cultured cells. Cells from coral tissue and those present in the cultures were found to be very similar (Fig. 3 A–C).
Sinularia flexibilis. Histological sections with ×40 magnification (A–C); (A) arrowed a coral cell with its endosymbiont. (B) Coral cells with one (a), two (b), or three (c) endosymbionts right after mechanical dissociation process. (C) Coral cells with zooxanthellae after 26 d of incubation in M-GIM.
Genetic test Using internal transcribed spacer primers.
To confirm the similarity of the cultured cells and those of the coral tissue, a genetic test using ITS was used. The conserved ribosomal genes (rDNA) in eukaryotes are separated by both highly variable external and internal TSs, one of which is the ITS used in this study. These typically include the 18S, 5.8S, and 25–28S ribosomal RNAs. Primers designed for amplification of ITS regions will anneal to more conserved regions, but more specific primers can be defined, which amplify only specific groups of organisms (LaJeunesse 2001). We used the primers 1s/2ss, designed for the genus Alcyonium (McFadden et al. 2001), closely related to Sinularia. These primers flank a region of approximately 1,400 bp of the nuclear ribosomal gene complex spanning the 3′ end of the 18 S subunit, ITS-1, 5.8 S subunit, ITS-2. If PCR products were obtained from the cell culture DNA extraction and from the coral tissue using these specific primers and if these had the same size, the presence of coral and zooxanthellae cells will be confirmed.
Using this primer set 1s/2ss (McFadden et al. 2001), a fragment of approximately 1,400 base pairs was obtained using PCR. The band was observed for all the undiluted samples, confirming the coral cells in the various cultures and in the intact animal to be identical. For the dilutions, in contrast, only the samples from the coral tissue presented a band for all of them and, as it is seen in Fig. 4, the higher the dilution, the lower its intensity; this may be explained by the lower amount of template DNA available for the PCR reaction in the dilutions. For GMIM, culture bands were visible up to 100 times diluted, for GIM culture up to ten times diluted, and for DMEM culture only a slight band for the undiluted sample (Fig. 4).
Electrophoresis gel showing 1s/2ss primer combination. A band of approximately 1400 bp is visible for all the undiluted samples (1), confirmation of the presence of the same kind of coral cells in all the different cultures and the coral tissue sample. M = Marker, 1 = undiluted sample, 2 = ten times diluted, 3 = 100 times diluted, 4 = 1,000 times diluted.
Those results are in line with the cellular growth rates (Fig. 1), where the higher cellular growth rates were obtained in GMIM medium and, to a lesser extent, in GIM medium. During the mentioned work, the medium DMEM did not show signs of growth, but the cells were preserved in good conditions.
Using the primers ITSa/ITSb specifically designed for volvocacean green algae (Coleman et al. 1994), also the region of the nuclear ribosomal gene complex spanning the 3′ end of the 18S subunit, ITS-1, 5.8S subunit, ITS-2, and the 5′ end of the 28S subunit was amplified, a band of approximately 700 bp was observed for all the undiluted samples, demonstrating once more the presence of the same kind of symbiont cells in all the cultures and coral tissue (Fig. 5). Also, the algae symbiont (zooxanthellae) belongs most likely to the genus Symbiodinium (Loram et al. 2007).
Electrophoresis gel showing ITSa/ITSb primer combination. A band of approximately 700 bp are visible for all the undiluted samples (1), corroborating the presence of the same kind of symbionts in all the different cultures and the coral tissue sample. M = Marker, 1 = undiluted sample, 2 = ten times diluted, 3 = 100 times diluted, 4 = 1,000 times diluted, B = Blank.
As it was explained previously, the different dilutions also had an effect on the PCR reaction which is reflected in the band intensity (Figs. 4 and 5). Through this molecular test, the identity of the cultivated cells was established: they were found to be identical to native coral cells in all cases.
Conclusions
Mechanical dissociation of coral tissue for developing a primary coral cell culture was most successful. It yielded the highest number of extracted cells and allowed cell growth after subsequent cultivation. The best chemical dissociation agent was trypsin-EDTA. Coral cells obtained after spontaneous dissociation did not grow.
Due to their small area and easy handling, 24-well plates were found appropriate for the establishment of the first cultures. The antibiotics were found to be effective in disinfecting in 90% of the cultures. Light was a fundamental parameter for coral cell culture. The best growth of coral cells was obtained with the media GIM and GMIM. Although both DMEM and M199 preserved the cells in good conditions, no cell growth was observed.
The software Image-proplus was a helpful tool for the growth determination process, increasing the accuracy of the counts and partially reducing the time needed. The genetic test using ITS primers was a feasible technique to identify the similarity of the cultured cells and the original cells in the coral. Hence, it is applicable to future cell culture studies. Finally, the culture of coral cells seems to be possible; however, further investigations are needed in order to establish the optimum conditions for the culture development.
References
Ata A.; Kerr R.; Moya C.; Jacobs R. Identification of anti-inflammatory diterpenes from the marine gorgonian Pseudopterogorgia elisabethae. Tetrahedron 59: 4215–4222; 2003.
Chao C.; Hsieh C.; Chen S.; Lu C.; Daid C.; Sheua J. Sinularianins A and B, novel sesquiterpenoids from the Formosan soft coral Sinularia sp. Tetrahedron Letters 47: 5889–5891; 2006.
Coleman A.; Suarez A.; Goff L. Molecular delineation of species and syngens in volvocacean green algae (Chlorophyta). Journal of Phycology 30: 80–90; 1994.
Domart-Coulon I.; Elbert D.; Scully E.; Calimlim P.; Ostrander G. Aragonite crystallization in primary cell cultures of multicellular isolates from a hard coral Pocillopora damicornis. PNAS 98: 11885–11890; 2001.
Domart-Coulon J.; Sinclair C.; Hill R.; Tambutté S.; Puverel S.; Ostrander G. A basidiomycete isolated from the skeleton of Pocillopora damicornis (Scleractinia) selectively stimulates short-term survival of coral skeletogenic cells. Marine Biology 144: 583–592; 2004.
Duh C.; Wang S.; Tseng H.; Sheu J.; Chiang M. Novel Cytotoxic Cembranoids from the Soft Coral Sinularia flexibilis. Journal of Natural Products 61: 844–847; 1998.
Duque C.; Puyana M.; Narváez G.; Osorno O.; Hara N.; Fujimoto Y. Pseudopterosins P–V, new compounds from the gorgonian octocoral Pseudopterogorgia elisabethae from Providencia island, Colombian Caribbean. Tetrahedron 60: 10627–10635; 2004.
Faith M.; Tachibana S.; Chou L.; Ding J. Citotoxic and anticancer agents in mucus of Galaxea fascicularis purification and characterization. Hournal of Marine Biotechnology 5: 50–57; 1997.
Frank U.; Rinkevich B. Scyphozoan jellyfish’s mesoglea supports attachment, spreading and migration of anthozoans’ cells in vitro. Cell Biology International 23: 307–311; 1999.
Frank U.; Rabinowitz C.; Rinkevich B. In vitro establishment if continuous cell cultures and cell lines from ten colonial cnidarians. Marine Biology 120: 491–499; 1994.
Freshney R. Culture of animal cells: a manual of basic technique. 3rd ed. Wiley, New York1994.
Gates R.; Baghdasarian B.; Muscatine L. Temperature stress causes host cell detachment in symbiotic cnidarians: Implications for coral bleaching. Biological Bulletin 182: 324–332; 1992.
Hunt B.; Vincent A. Scale and sustainability of marine bioprospecting for pharmaceuticals. Ambio 352: 57–64; 2006.
Hurton L.; Berkcson J.; Smith S. Selection of a standard culture media for primary culture of Limulus polyphemus amebocytes. In vitro cell. Developmental Biology 41: 325–329; 2005.
Kamel H.; Slattery M. Terpenoids of Sinularia: chemistry and biomedical applications. Pharmaceutical Biology 433: 253–269; 2005.
Kingsley R. J.; Bernhardt A. M.; Wilbur K. M.; Watabe N. Scleroblast cultures from the gorgonian leptogorgia virgulata (Lamarck} (Coelenterata: Gorgonacea}. In Vitro Cellular and Developmental Biology 234: 297–302; 1987.
Kopecky E.; Ostrander G. Isolation and primary culture of viable multicellular endothelial isolates from hard corals. In vitro Cellular and Developmental Biology. Animal 35: 616–624; 1999.
LaJeunesse T. Investigating the biodiversity, ecology, and phylogeny of endosymbiotic dinoflagellates in the genus Symbiodinium using the ITS region in search of a “species” level marker. Journal of Phycology 37: 866–880; 2001.
Loram J.; Boonham N.; O’toole P.; Trapido-Rosenthal H.; Douglas A. Molecular quantification of symbiotic dinoflagellate algae of the genus Symbiodinium. Biological Bulletin 212: 259–268; 2007.
McFadden C.; Donahue R.; Hadland B.; Weston R. A molecular phylogenetic analysis of reproductive trait evolution in the soft coral genus Alcyonium. Evolution 551: 54–67; 2001.
Odintsova N. A.; Ermak A. V.; Tsal L. G. Substrate selection for long-term cultivation of marine invertebrate cells. Camp. Biochem. Physiol 1074: 613–619; 1994.
Pomponi S. A.; Willoughby R.; Kaighn M. E.; Wright A. E. Development of techniques for in vitro production of bioactive natural products from marine sponges. In: MaramoroschK.; MitsuhashiJ. (eds) Invertebrate cell culture: novel directions and biotechnology applications. Science, Enfield, pp 231–238; 1997.
Puverel S.; Tambutte´ E.; Zoccola D.; Domart-Coulon I.; Bouchot A.; Lotto S. et al. Antibodies against the organic matrix in scleractinians: a new tool to study coral biomineralization. Coral Reefs 24: 149–156; 2005.
Radhika P.; Rajeswara R.; Archana J.; Koteswara N. Anti-inflammatory activity of a new sphingosine derivative and cembrenoid diterpene (Lobohedleolide) isolated from marine soft corals of Sinularia crassa Tixier-Durivault and Lobophytum species of the Andaman and Nicobar Islands. Biological & Pharmaceutical Bulletin 287: 1311–1313; 2005.
Rinkevich B. Cell cultures from marine invertebrates: obstacles, new approaches and recent improvements. Journal of Biotechnology 70: 133–153; 1999.
Rinkevich B. Marine invertebrate cell cultures: new millennium trends. Marine Biotechnology 7: 429–439; 2005.
Roche-applied Science. Pronase Nuclease-free, isolated from Streptomyces griseus Lyophilized powder. Accessed 2007. http://www.roche-applied-science.com/pack-insert/0165921a.pdf; 2006.
Rodgers C. J. Book review. In: Mothersill C.; AustinB. (eds) Aquatic invertebrate cell culture. Springer in association with Praxis, Chichester2002.
Schmid V.; Ono S.; Reber-Müller S. Cell-substrate interaction in Cnidaria. Microscopy Research and Technique 44: 254–268; 1999.
Sigma-Aldrich. Typsine-EDTA solution cell culture tested. Accessed 2007. http://www.roche-applied-science.com/pack-insert/0165921a.pdf; 2007.
Smith D. Symbiosis research at the end of the millennium. Hydrobiologia 461: 49–54; 2001.
Verkerk M.; van den End F.; Martens D. Animal cell technology (PRE 30806) practical guide. Food and bioprocess engineering group. Wageningen University, Wageningen 2007.
Villena A. J. Applications and needs of fish and shellfish cell culture for disease control in aquaculture. Reviews in Fish Biology and Fisheries 13: 111–140; 2003.
Yu S.; Deng Z.; van Ofwegen L.; Proksch P.; Lin W. 5,8-Epidioxysterols and related derivatives from a Chinese Soft Coral Sinularia flexibilis. Steroids 71: 955–959; 2006.
Acknowledgements
This study was funded by the government of I.R. of Iran. We are grateful to Anja Taverne-Thiele and Nico Taverne from Animal Sciences department (Wageningen University) for their collaboration with the histological slides; and to Boudewijn van Veen form the Microspectroscopy Centre (Wageningen University) for his guide with the image software.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Editor: J. Denry Sato
An erratum to this article can be found at http://dx.doi.org/10.1007/s11626-008-9150-9
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Khalesi, M.K. Cell cultures from the symbiotic soft coral Sinularia flexibilis . In Vitro Cell.Dev.Biol.-Animal 44, 330–338 (2008). https://doi.org/10.1007/s11626-008-9128-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11626-008-9128-7