Abstract
Objective
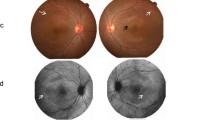
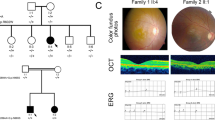
Autosomal recessive bestrophinopathy (ARB), a retinal degenerative disease, is characterized by central visual loss, yellowish multifocal diffuse subretinal deposits, and a dramatic decrease in the light peak on electrooculogram. The potential pathogenic mechanism involves mutations in the BEST1 gene, which encodes Ca2+-activated Cl− channels in the retinal pigment epithelium (RPE), resulting in degeneration of RPE and photoreceptor. In this study, the complete clinical characteristics of two Chinese ARB families were summarized.
Methods
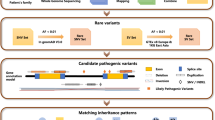
Pacific Biosciences (PacBio) single-molecule real-time (SMRT) sequencing was performed on the probands to screen for disease-causing gene mutations, and Sanger sequencing was applied to validate variants in the patients and their family members.
Results
Two novel mutations, c.202T>C (chr11:61722628, p.Y68H) and c.867+97G>A, in the BEST1 gene were identified in the two Chinese ARB families. The novel missense mutation BEST1 c.202T>C (p.Y68H) resulted in the substitution of tyrosine with histidine in the N-terminal region of transmembrane domain 2 of bestrophin-1. Another novel variant, BEST1 c.867+97G>A (chr11:61725867), located in intron 7, might be considered a regulatory variant that changes allele-specific binding affinity based on motifs of important transcriptional regulators.
Conclusion
Our findings represent the first use of third-generation sequencing (TGS) to identify novel BEST1 mutations in patients with ARB, indicating that TGS can be a more accurate and efficient tool for identifying mutations in specific genes. The novel variants identified further broaden the mutation spectrum of BEST1 in the Chinese population.
Similar content being viewed by others
References
Marquardt A, Stöhr H, Passmore LA, et al. Mutations in a novel gene, VMD2, encoding a protein of unknown properties cause juvenile-onset vitelliform macular dystrophy (Best’s disease). Hum Mol Genet, 1998,7(9):1517–1525
Marmorstein AD, Marmorstein LY, Rayborn M, et al. Bestrophin, the product of the Best vitelliform macular dystrophy gene (VMD2), localizes to the basolateral plasma membrane of the retinal pigment epithelium. Proc Natl Acad Sci USA, 2000,97(23):12758–12763
Petrukhin K, Koisti MJ, Bakall B, et al. Identification of the gene responsible for Best macular dystrophy. Nat Genet, 1998,19(3):241–247
Burgess R, Millar ID, Leroy BP, et al. Biallelic mutation of BEST1 causes a distinct retinopathy in humans. Am J Hum Genet, 2008,82(1):19–31
Allikmets R, Seddon JM, Bernstein PS, et al. Evaluation of the Best disease gene in patients with age-related macular degeneration and other maculopathies. Hum Genet, 1999,104(6):449–453
Yardley J, Leroy BP, Hart-Holden N, et al. Mutations of VMD2 splicing regulators cause nanophthalmos and autosomal dominant vitreoretinochoroidopathy (ADVIRC). Invest Ophthalmol Vis Sci, 2004,45(10):3683–3689
Davidson AE, Millar ID, Urquhart JE, et al. Missense mutations in a retinal pigment epithelium protein, bestrophin-1, cause retinitis pigmentosa. Am J Hum Genet, 2009,85(5):581–592
Habibi I, Falfoul Y, Todorova MG, et al. Clinical and Genetic Findings of Autosomal Recessive Bestrophinopathy (ARB). Genes (Basel), 2019,10(12):953
Poplin R, Chang PC, Alexander D, et al. A universal SNP and small-indel variant caller using deep neural networks. Nat Biotechnol, 2018,36(10):983–987
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res, 2010,38(16):e164
Tian R, Yang G, Wang J, et al. Screening for BEST1 gene mutations in Chinese patients with bestrophinopathy. Mol Vis, 2014,20:1594–1604
Petersen BS, Fredrich B, Hoeppner MP, et al. Opportunities and challenges of whole-genome and -exome sequencing. BMC Genet, 2017,18(1):14
Salzberg SL, Yorke JA. Beware of mis-assembled genomes. Bioinformatics, 2005,21(24):4320–4321
Guan P, Sung WK. Structural variation detection using next-generation sequencing data: A comparative technical review. Methods, 2016,102:36–49
Bayega A, Wang YC, Oikonomopoulos S, et al. Transcript Profiling Using Long-Read Sequencing Technologies. Methods Mol Biol, 2018,1783:121–147
Steijger T, Abril JF, Engström PG, et al. Assessment of transcript reconstruction methods for RNA-seq. Nat Methods, 2013,10(12):1177–1184
Conlin LK, Aref-Eshghi E, McEldrew DA, et al. Long-read sequencing for molecular diagnostics in constitutional genetic disorders. Hum Mutat, 2022,43(11):1531–1544
Milenkovic VM, Rivera A, Horling F, et al. Insertion and topology of normal and mutant bestrophin-1 in the endoplasmic reticulum membrane. J Biol Chem, 2007,282(2):1313–1321
Milenkovic A, Milenkovic VM, Wetzel CH, et al. BEST1 protein stability and degradation pathways differ between autosomal dominant Best disease and autosomal recessive bestrophinopathy accounting for the distinct retinal phenotypes. Hum Mol Genet, 2018,27(9):1630–1641
Marmorstein AD, Kinnick TR, Stanton JB, et al. Bestrophin-1 influences transepithelial electrical properties and Ca2+ signaling in human retinal pigment epithelium. Mol Vis, 2015,21:347–359
Rosenthal R, Bakall B, Kinnick T, et al. Expression of bestrophin-1, the product of the VMD2 gene, modulates voltage-dependent Ca2+ channels in retinal pigment epithelial cells. FASEB J, 2006,20(1):178–180
Strauß O, Müller C, Reichhart N, et al. The role of bestrophin-1 in intracellular Ca(2+) signaling. Adv Exp Med Biol, 2014,801:113–119
Gao FJ, Qi YH, Hu FY, et al. Mutation spectrum of the bestrophin-1 gene in a large Chinese cohort with bestrophinopathy. Br J Ophthalmol, 2020,104(6):846–851
Qu Z, Hartzell C. Determinants of anion permeation in the second transmembrane domain of the mouse bestrophin-2 chloride channel. J Gen Physiol, 2004,124(4):371–382
Uggenti C, Briant K, Streit AK, et al. Restoration of mutant bestrophin-1 expression, localisation and function in a polarised epithelial cell model. Dis Model Mech, 2016,9(11):1317–1328
Davidson AE, Millar ID, Burgess-Mullan R, et al. Functional characterization of bestrophin-1 missense mutations associated with autosomal recessive bestrophinopathy. Invest Ophthalmol Vis Sci, 2011,52(6):3730–3736
Johnson AA, Bachman LA, Gilles BJ, et al. Autosomal Recessive Bestrophinopathy Is Not Associated With the Loss of Bestrophin-1 Anion Channel Function in a Patient With a Novel BEST1 Mutation. Invest Ophthalmol Vis Sci, 2015,56(8):4619–4630
Johnson AA, Lee YS, Chadburn AJ, et al. Disease-causing mutations associated with four bestrophinopathies exhibit disparate effects on the localization, but not the oligomerization, of Bestrophin-1. Exp Eye Res, 2014,121:74–85
Kinnick TR, Mullins RF, Dev S, et al. Autosomal recessive vitelliform macular dystrophy in a large cohort of vitelliform macular dystrophy patients. Retina, 2011,31(3):581–595
Gerth C, Zawadzki RJ, Werner JS, et al. Detailed analysis of retinal function and morphology in a patient with autosomal recessive bestrophinopathy (ARB). Doc Ophthalmol, 2009,118(3):239–246
Boon CJ, van den Born LI, Visser L, et al. Autosomal recessive bestrophinopathy: differential diagnosis and treatment options. Ophthalmology, 2013,120(4):809–820
Xiao Q, Hartzell HC, Yu K. Bestrophins and retinopathies. Pflugers Arch, 2010,460(2):559–569
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that there are no conflicts of interest with any financial organization, corporation or individual that can inappropriately influence this work.
Rights and permissions
About this article
Cite this article
Li, Jx., Meng, Lr., Hou, Bk. et al. Detection of Novel BEST1 Variations in Autosomal Recessive Bestrophinopathy Using Third-generation Sequencing. CURR MED SCI 44, 419–425 (2024). https://doi.org/10.1007/s11596-024-2865-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11596-024-2865-3